Abstract
Accumulating evidence hints heterochromatin anchoring to the inner nuclear membrane as an upstream regulatory process of gene expression. Given that the formation of neural progenitor cell lineages and the subsequent maintenance of postmitotic neuronal cell identity critically rely on transcriptional regulation, it seems possible that the development of neuronal cells is influenced by cell type-specific and/or context-dependent programmed regulation of heterochromatin anchoring. Here, we explored this possibility by genetically disrupting the evolutionarily conserved barrier-to-autointegration factor (Baf) in the Drosophila nervous system. Through single-cell RNA sequencing, we demonstrated that Baf knockdown induces prominent transcriptomic changes, particularly in type I neuroblasts. Among the differentially expressed genes, our genetic analyses identified teashirt (tsh), a transcription factor that interacts with beta-catenin, to be closely associated with Baf knockdown-induced phenotypes that were suppressed by the overexpression of tsh or beta-catenin. We also found that Baf and tsh colocalized in a region adjacent to heterochromatin in type I NBs. Notably, the subnuclear localization pattern remained unchanged when one of these two proteins was knocked down, indicating that both proteins contribute to the anchoring of heterochromatin to the inner nuclear membrane. Overall, this study reveals that the Baf-mediated transcriptional regulation of teashirt is a novel molecular mechanism that regulates the development of neural progenitor cell lineages.

Similar content being viewed by others
Introduction
The nervous system consists of diverse cellular components including various types of neurons and glial cells. The complex cellular composition of the nervous system requires the tightly controlled generation of specific cell types from neural progenitor cells during development, which involves asymmetric/symmetric cell division and spatiotemporal expression of transcription factors (TFs) that determine cell identity. Extensive studies have been conducted to identify factors regulating neurodevelopment and revealed that the orchestrated action of TFs is crucial for both the formation of neural progenitor cell lineages and the subsequent maintenance of postmitotic neuronal cell identity1,2,3. For instance, neural progenitors determine the cellular identity of their progenies4,5 through the sequential upregulation of temporal TFs, such as Hunchback (Hb), Kruppel (Kr), POU domain protein (Pdm), and Castor (Cas), which was first characterized in embryonic neuroblasts (NBs) of the Drosophila ventral nerve cord6,7. In addition, regarding the maintenance of postmitotic neuronal cell identity, the initiation and subsequent maintenance of olfactory receptor expression patterns in Drosophila olfactory sensory neurons are regulated by combinations of seven TFs (acj6, E93, Fer1, onecut, sim, xbp1, and zf30c)8. However, the upstream activators/cues or regulatory processes that control the activities and/or amounts of these TFs in cell type-specific and/or context-dependent (e.g., developmental stages or cell cycle phases) manners during nervous system development remain largely unknown.
In this regard, it is worth noting that recent studies have highlighted the roles of chromosomal features (i.e., higher-order chromatin organization and subnuclear positioning of chromatin) in the regulation of gene expression9,10. As for the nervous system, the roles of chromatin organization in neurodevelopment, specifically via transcriptional regulation, have recently been actively explored11,12,13,14, whereas the roles of subnuclear positioning of chromatin involving transcriptional regulation have remained relatively less understood. The anchoring of heterochromatin to the inner nuclear membrane (INM) is crucial for subnuclear positioning of chromatin15,16, which is mediated by chromatin-binding proteins/chromatin-anchoring proteins, such as the evolutionally conserved barrier-to-autointegration factor (Baf)17,18,19, mammalian proline-rich-protein (PRR14)20, and C. elegans-specific chromodomain-containing protein (cec-4)21,22. These chromatin-binding proteins/chromatin-anchoring proteins interact with lamin-associated proteins, such as proteins with LEM (LAP2, Emerin, MAN1) domains that reside in the INM23,24,25, to form physical interactions between the chromosome and the nuclear lamina. According to this anchoring function, investigations on the anchoring of heterochromatin to the INM have primarily focused on its involvement in chromosomal segregation and subsequent postmitotic reassembly of the nuclear envelope during mitosis26,27,28.
Beyond their well-characterized roles in mitosis, chromatin-binding proteins/chromatin-anchoring proteins can play a role in regulating gene expression. For instance, Baf is associated with the transcriptional regulation of target genes, which involves its direct interactions with promoter regions, recruitment of epigenetic modulators, and/or histone modifications17,18,29. Furthermore, accumulating evidence suggests the potential importance of heterochromatin anchoring to the INM in the differentiation of cells into specific cell types during development. A previous study reported that baf-1, a C. elegans ortholog of Baf, plays a postmitotic role in muscle and epidermal cells; homozygous baf-1 mutants are able to grow into L4 larvae/young adults but exhibit cell type-specific defects, such as defects in muscle cell integrity and premature fusion of epidermal cells, accompanied by a reduced body size29. Moreover, PRR14 is associated with myoblast differentiation that involves transcriptional regulation of specific target genes (e.g., MyoD) through its interaction with HP1alpha30. In this respect, we here investigated whether heterochromatin anchoring-dependent regulation of transcription affects neurodevelopment in a cell type-specific and/or context-dependent manner using Drosophila as a model. To this end, we specifically focused on the evolutionally conserved Baf31, since there is no known fly homolog of PRR14 or cec-4. To genetically disrupt the programmed regulation of heterochromatin anchoring during nervous system development, we used Baf-knockdown (KD) flies instead of Baf-null mutant flies because complete knockout of Baf results in mortality during the larval–pupal transition32. By combining single-cell transcriptomic analysis with a series of genetic analyses, we found that Baf-mediated transcriptional regulation of a specific downstream molecule, teashirt (tsh), which interacts with beta-catenin in the Wnt signaling pathway, is involved in the Baf-dependent regulation of heterochromatin anchoring to the INM, primarily in type I NBs, and the subsequent formation of neural progenitor cell lineages in Drosophila.
Materials and methods
Fly stocks
The following fly stocks were obtained from the Bloomington Drosophila Stock Center (Indiana, USA): elav-Gal4 (BL8760), nSyb-Gal4 (BL51941), repo-Gal4 (BL7415), UAS-Baf RNAi (BL36108), UAS-Baf RNAi (KK102013), OK371-QF2 (BL66473), QUAS-mCD8GFP (BL30003), UAS-Redstinger (BL8546), wor-Gal4,UAS-mira.GFP/CyO, ActGFP;UAS-His-RFP (BL56555), neoFRT40A (BL8212), l(2)k10210 (BL10980), UAS-tsh RNAi (BL35030), UAS-msk RNAi (BL33626), UAS-mts RNAi (BL38337), UAS-CG5033 RNAi (BL42930), UAS-Spindly RNAi (BL34933), UAS-Uba2 RNAi (BL63986), UAS-mars RNAi (BL33929), UAS-U4-U6-60K RNAi (BL55210), UAS-Caf1-55 RNAi (BL34069), UAS-blw RNAi (BL28059), UAS-vvl RNAi (BL50657), UAS-feo RNAi (BL28926), UAS-Hsp70Bc RNAi (BL42626), UAS-Rap1 RNAi (BL35047), UAS-puf RNAi (BL62477), UAS-foxo RNAi (BL32993), UAS-ZnT49B RNAi (BL31933), UAS-Ranbp9 RNAi (BL33004), UAS-CG43340 RNAi (BL41926), UAS-tsh (BL52216), UAS-tsh (BL52217), UAS-msk (BL23944), UAS-Arm (BL8369), UAS-Arm.S10 (BL4782), and UAS-Arm RNAi (BL35004). UAS-OdsH-3xHA (F000261), UAS-FoxK-3xHA (F000615), and UAS-msk-3xHA (F004312) were obtained from FlyORF (Zurich, Switzerland). Type I NB-Gal4 (ase-Gal4) and type II NB-Gal4 (wor-Gal4, ase-Gal80) were kindly provided by J.A. Knoblich (IMBA, Austria). Tub-Gal80TS, UAS-mCD8GFP, and UAS-mCD4GFP were gifts from Y.N. Jan (University of California, San Francisco, CA). elav-Gal4, UAS-mCD8GFP, hsflp;FRT40A, and Tub-Gal80 were kindly provided by J. Chung (Seoul National University, Korea). All flies were raised at room temperature (25 °C) with 60% humidity.
Between two available transgenic lines for overexpression of tsh, we observed that overexpression of tsh in type I NBs using the BL52216 transgenic fly line, but not the BL52217 transgenic fly line, induces early lethality during the first instar larval stage. Thus, the genetic interaction between Baf and tsh was first examined in the BL52217 line and subsequently confirmed in the BL52216 line co-expressed with Gal80 in the parents to prevent the early death of the progeny through the maternal supply of Gal80, or vice versa.
Culture of primary NBs for live imaging
Culture of primary NBs was performed as previously described33. Third-instar larvae were briefly washed with PBS. Larval brains were dissected in Dissection medium [Schneider’s medium (Sigma) supplemented with 10% fetal bovine serum and 2% Pen/Strep] and collected in cold Rinaldini solution (800 mg NaCl, 20 mg KCl, 5 mg NaH2PO4, 100 mg NaHCO3, and 100 mg glucose in 100 ml distilled water). After brief washing with Rinaldini solution, the brains were incubated in Dissociation medium [Rinaldini solution containing 0.5 mg/ml collagenase I and 1 mg/ml papain (Sigma)] for 1 h at 30 °C to dissociate the tissues into individual cells. After 1 h, the dissociated cells were washed with Rinaldini solution and then with Dissection medium. The dissociated cells were subsequently resuspended in 200 μl of Dissection medium and plated in poly-L-lysin-coated cell culture dishes (FD35-100, FluoroDish). The culture dishes were incubated in a 25 °C incubator for 1 h, after which 3 ml of primary cell culture medium [Schneider’s medium (Sigma) supplemented with 10% fetal bovine serum, 2% Pen/Strep, 20 mM L-glutamine, 5 μg/ml L-glutathione, 20 μg/ml insulin, and 5 μg/ml ecdysone] was added before imaging.
Culture of primary NBs for immunocytochemistry
Culture of primary NBs was conducted as previously described34. Third-instar larvae were rinsed with 1x PBS. All the solutions that were used for primary culture were kept cold on ice. Larval brains were dissected in Dissection medium [90% L-glutamine-supplemented Schneider’s medium, 10% heat-inactivated FBS, 0.1% penicillin/streptomycin]. Using type I NBs localized in the central lobe of the brain, we removed ventral nerve cords from each dissected larval brain and collected a total of 5 pairs of third-instar larval brain lobes in ice-cold Rinaldini solution [800 mg NaCl, 20 mg KCl, 5 mg NaH2PO4H2O, 100 mg NaHCO3, 100 mg glucose, in 100 ml distilled water]. After brief washing with Rinaldini solution, the larval brain lobes were incubated in Dissociation medium [Rinaldini solution containing 0.5 mg/ml collagenase I] for 1 h at room temperature, followed by washing 3 times with Rinaldini solution. Then, 10 µl of Dissection medium was added to each of the larval brain samples. Subsequently, the brain lobes were homogenized by gentle trituration using a micropipette. Dissociated cells were seeded on the 0.01% poly-L-lysine-coated coverslip and incubated in a wet chamber for 1 h in room temperature to allow the dissociated cells to settle. To maintain the humidity inside the wet chamber, we used a microcentrifuge tube storage box and placed a small bucket of water on the bottom of the box. The cells were fixed with 3.7% formaldehyde for 15 min at room temperature and briefly washed three times every 2 min using 0.3% PBT (0.3% Triton X-100 in phosphate-buffered saline (PBS)). Then, the samples were blocked with 10% NDS containing 0.3% PBT for 1 h at room temperature and then incubated with primary antibodies diluted in 5% NDS containing 0.3% PBT overnight at 4 °C. After overnight incubation, the samples were washed three times every 10 min with 0.3% PBT containing Hoechst (1:200 dilution). The coverslips were mounted in ProLongTM Diamond Antifade Mountant for confocal imaging.
Immunohistochemistry and immunocytochemistry
For immunohistochemistry, larval or adult brains were dissected in 1x PBS and stained as previously described35. The following primary antibodies were used: rat anti-Dpn (ab195173, Abcam; 1:100 dilution); mouse anti-BrdU (347580, BD Biosciences; 1:250 dilution); rabbit anti-GFP (ab183734, Abcam; 1:100 dilution); mouse anti-GFP (ab11120, Abcam; 1:200 dilution); mouse anti-Pros (MR1A, Developmental Studies Hybridoma Bank; 1:10 dilution); rat anti-Miranda (ab197788, Abcam; 1:100 dilution); rabbit anti-PH3 (9701, Cell Signaling Technology, 1:100); rabbit anti-TH (AB152, MERCK; 1:200 dilution); mouse anti-ChAT (ChAT4B1, Developmental Studies Hybridoma Bank; 1:10 dilution); rabbit anti-5-HT (20080, IMMUNOSTAR; 1:500 dilution); and rabbit anti-GABA (A2052, Sigma-Aldrich; 1:100 dilution).
To detect these primary antibodies, the following secondary antibodies were used: Alexa 488-conjugated goat anti-rabbit (Thermo Fisher Scientific; 1:600 dilution), Alexa 555-conjugated goat anti-rat (Thermo Fisher Scientific; 1:600 dilution), Cy3-conjugated donkey anti-mouse (Jackson ImmunoResearch Laboratories; 1:200 dilution), and Alexa 647-conjugated goat anti-rat (Jackson ImmunoResearch Laboratories; 1:600 dilution). Images of immunostained brain samples were captured using Zeiss confocal microscopes.
For BrdU labeling of NBs, dissected larval brains were incubated in 40 ng/ml BrdU in Schneider medium for 30 min at room temperature and subsequently transferred to Schneider medium and incubated for 24 h at 4 °C. After incubation, the larval brains were washed three times with 0.3% PBT and fixed with 3.7% formaldehyde in 0.3% PBT for 30 min at room temperature. Next, the larval brains were exposed to 1 N HCl for 30 min and washed three times with 0.3% PBT. Then, the samples were stained as previously described35.
For Hoechst staining, dissected larval brains were incubated in 0.3% PBT containing 5 μg/ml Hoechst for 40 min at room temperature. After brief washing with 0.3% PBT, the larval brains were mounted in PBS prior to imaging.
The following primary antibodies were used for immunocytochemistry: rat anti-Dpn (ab195173; Abcam; 1:50 dilution), rabbit anti-H3K9me3 (ab8898; Abcam; 1:100 dilution), mouse anti-Arm (N2 7A1; Developmental Studies Hybridoma Bank; 1:100 dilution), and mouse anti-GFP (ab11120; Abcam; 1:100 dilution). Rabbit anti-Baf was kindly provided by Fernando Azorin (Institute of Molecular Biology of Barcelona, CSIC, Barcelona, Spain). Guinea pig anti-tsh antibody was a gift from Richard S. Mann (Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY). To detect these primary antibodies, the following secondary antibodies were used: Alexa 555-conjugated goat anti-rat (Thermo Fisher Scientific; 1:400 dilution), Alexa 647-conjguated goat anti-rabbit (Thermo Fisher Scientific; 1:400 dilution), Alexa 488-conjugated goat anti-mouse (Thermo Fisher Scientific; 1:400 dilution), Alexa 647-conjugated goat anti-guinea pig (Jackson ImmunoResearch Laboratories; 1:400 dilution), and Hoechst 34580 (Thermo Fisher Scientific; 1:200 dilution).
RT‒PCR
RT‒PCR analysis was performed as previously described36. Total RNA was extracted from adult fly heads using an Easy-Blue system (iNtRON Biotechnology). cDNAs were synthesized from 3 µg of total RNA using GoScript Reverse Transcription (A2791; Promega) following the manufacturer’s standard protocol. For RT‒PCR analysis, each target gene was amplified with the corresponding primer set (Supplementary Table 1) using GoTaq G2 Master Mix (M7823; Promega) in a C1000 Thermal Cycler, C1000 Touch Thermal Cycler, or T100 Thermal Cycler system (Bio-Rad).
Live imaging of cultured primary NBs
To monitor the cell division of NBs, live imaging of cultured primary NBs was performed using an LSM7 live confocal microscope (equipped with an imaging chamber, 25 °C) with a 40x water/1.2 NA objective. The cell division of the NBs was recorded for 3 h, and Z-stack images were acquired every 180 s at 1 μm intervals.
Calculation of the circularity of NBs
The circularity of NBs was calculated as previously described26. Confocal images were processed to determine the maximum projection intensity, and the perimeter (P) and area (A) of the NBs were measured using ImageJ. The circularity of the NBs was subsequently calculated using the following formula: \(P/(2\,\cdot\, \sqrt{\pi \,\cdot\, A})\).
Sample preparation for single-cell RNA sequencing (scRNA-seq)
For single-cell sequencing, third-instar larvae were collected and prepared as previously described33. Third-instar larvae expressing denoted transgenes (elav-Gal4 and elav-Gal4/UAS-Baf RNAi) were dissected in 1x PBS and collected in cold 1x PBS. The collected larval brains were centrifuged at 800 × g for 5 min, after which the supernatants were replaced with dissociation buffer (0.6 mg/ml dispase, 0.15 mg/ml collagenase I, and 0.025% trypsin-EDTA). The larval brains were incubated in a thermoshaker (25 °C) for 15 min at 1000 rpm. The dissociated brain cells were washed with cold 1x PBS and resuspended in Dulbecco’s phosphate-buffered saline (DPBS) containing 0.04% bovine serum albumin. The dissociated brain cells were subsequently filtered through a 40-μm strainer (Flowmi® Cell Strainers) and collected for the subsequent steps related to single-cell sequencing.
scRNA-seq
Third-instar larval brains were collected and prepared as previously described33. Briefly, third-instar larvae of the elav-Gal4 and elav-Gal4/UAS-Baf RNAi fly stocks were dissected in 1x PBS and collected in cold 1x PBS. The collected larval brains were centrifuged at 800 × g for 5 min, after which the supernatants were replaced with dissociation buffer (0.6 mg/ml dispase, 0.15 mg/ml collagenase I, and 0.025% trypsin-EDTA). The larval brains were incubated in a thermoshaker (25 °C) for 15 min at 1000 rpm. The dissociated brain cells were washed with cold 1x PBS and resuspended in Dulbecco’s phosphate-buffered saline (DPBS) containing 0.04% bovine serum albumin. These dissociated cells were filtered with a 40-μm strainer (Flowmi® Cell Strainers), fixed with cold 80% methanol and stored at −20 °C until all the replicates were collected. To rehydrate the samples, the cells were centrifuged at 3000 × g for 5 min at 4 °C, after which the supernatants were removed. The cell pellets were washed 3 times with 1% BSA and 1% Superase • In RNase inhibitor (Ambion) in DPBS. The cells were resuspended at a concentration of 1 × 106 cells/ml in cold 0.5x DPBS supplemented with an RNase inhibitor (Enzynomics).
The sequencing libraries for scRNA-seq were generated by SPLiT-seq37. Briefly, for in situ reverse transcription of the first-round barcode, cells were aliquoted into 48 wells of a 96-well plate, in which barcoded well-specific reverse transcription primers had been previously aliquoted. The second- and third-round barcodes were appended to the cDNA by an in-cell ligation step. After the third-round barcodes were appended, 8000 cells were aliquoted into each sublibrary and lysed. cDNA was purified and amplified using PCR. The quality and quantity of cDNA were monitored using a Bioanalyzer high sensitivity kit (Agilent). A total of 600 pg of cDNA was used for tagmentation, and the i5/i7 sample index was inserted by PCR. Purified libraries were sequenced on the Illumina HiSeq X platform (paired-end 150-bp reads) aiming for a depth of 50,000 read pairs per cell.
scRNA-seq data preprocessing
The raw fastq files were processed using the zUMIs pipeline (v2.5.4)38. To extract UMI and cell barcode (CB) information from paired-end reads, the following base definitions were used in the YAML file: cDNA (1–151) from read 1, CB (11–18, 49–56, and 87–94) from read 2, and UMI (1–10) from read 2. The cDNA sequences were subsequently mapped to the Drosophila genome (BDGP6.28) using the STAR (v2.7.4a) aligner39 with the BDGP6.28.99 GTF file. A gene-by-cell count matrix was generated with default parameters, and cells with less than 100 total UMI counts were removed because the expected number of cells across samples was approximately 8000 cells. To filter out low-quality cells, cells with a total log10-scaled UMI count less than 2.2 and cells with more than 5% of the UMIs assigned to mitochondrial genes were excluded, and the thresholds were determined by visually inspecting the outliers in the PCA plot on the quality control metrics using the calculateQCMetrics function of the scater (v1.14.0) R package40. To remove cell-specific biases, cells were clustered using the quickCluster function of the scran (v1.14.6) R package41 with default parameters, and cell-specific size factors were computed using the computeSumFactors function of the same package. The aggregated gene-by-cell count matrix across samples was normalized by dividing the raw UMI counts by cell-specific size factors. The normalized counts were then log2-transformed by adding a pseudocount of 1. We defined highly variable genes (HVGs) as the 1000 genes with the highest biological variability using the decomposeVar function of the scran package. All the cells were grouped into 14 clusters using the FindClusters function of the Seurat (v3.2.0) R package42 on the first 10 PCs of HVGs with a resolution = 0.8 and visualized with a two-dimensional UMAP plot using the RunUMAP function of the same package on the same 10 PCs. Two clusters (Clusters 7 and 12) expressing multiple cell type markers were considered putative doublets and removed. The remaining cells were regrouped into 13 clusters on the 20 PCs of 1500 HVGs using the same method described above. For the analysis of neuronal lineage cells, neuronal cells (glial cells were excluded) were reclustered and visualized using the 7 PCs of 1500 HVGs.
scRNA-seq data analysis
We identified differentially expressed genes (DEGs) in each cell type between the control and Baf KD groups using the limma (v3.44.3) R package43 with an adjusted P < 0.05 and an absolute value of log2FC > 0.1. Cell types responsive to Baf KD were prioritized based on the cross-validation area under the receiver operating characteristic curve (AUC) of the random forest classifier, which was implemented in the Augur (v1.0.0) R package with default parameters44. To determine the biological processes in which DEGs in each cell type between the control and Baf KD groups were enriched, significantly enriched GO biological process (GOBP) terms (P < 0.05) were selected using the topGO (v2.40.0) R package with the org.Dm.eg.db (v3.11.4) annotation data package. We performed pseudotime analysis of both the type I NB lineage (type I NB – GMC – neuron) and the type II NB lineage (type II NB – INP– GMC – neuron) under control or Baf KD conditions using the Palantir (v0.2.6) python package45. A k-nearest neighbor (kNN) graph (k = 30) was constructed using the first 10 diffusion components (DCs) derived from the 100 PCs of 1500 HVGs and visualized in the t-SNE plot based on the same 10 DCs. The starting cells for the pseudotime analysis were defined by choosing the cell with the highest expression of ase for the type I NB lineage and pnt for the type II NB lineage. A lineage tree in the same UMAP plot was generated for each lineage under each condition after separating cells according to their lineage and treatment condition. We constructed a kNN graph using the pp.neighbors function of the scanpy (v1.5.1) python package with 7 PCs. The connectivity of the cell types in each lineage under each condition was quantified using the tl.paga function of the same package with default parameters. The edge weights were visualized using the pl.paga function of the same package with the root partitions and layout Reingold-Tilford. To compare the neuronal development of each NB lineage between the control and Baf KD groups, we aligned two trajectories using the cellAlign (v0.1.0) R package46 with the Palantir pseudotime values of cells and 300 interpolated points for each trajectory.
Behavioral analyses of adult flies
After eclosion from pupae, adult male flies were collected in vials containing fresh food and reared (25 °C, 60% humidity, and 10 AM:10 PM light:dark cycle) for 3–4 days. Then, each fly was separately transferred to a single fresh vial without CO2 anesthesia 12 h before behavioral analyses. On the day of the analyses, each fly was individually transferred to a recording chamber without CO2 anesthesia for the behavioral analyses. After 3 min of acclimation, the behavior of each fly was recorded for 10 min.
Quantification and statistical analysis
For statistical comparisons of the quantified results between two different groups, two-tailed Student’s t-tests were used. When comparing three or more groups, one- or two-way ANOVA was used with Tukey’s post hoc correction. All these comparisons were performed using GraphPad Prism (version 7, GraphPad Software, Inc.). N.S., *, **, ***, and **** indicate P > 0.05, P < 0.05, P < 1.0 × 10−2, P < 1.0 × 10−3, and P < 1.0 × 10−4, respectively. All the data are shown as the mean ± SEM. The value of n represents the number of animals.
Results
scRNA-seq revealed that Baf KD induces prominent transcriptional changes, particularly in type I NBs
To understand the neurodevelopmental roles of Baf, we first characterized the cell type-specific transcriptional changes induced by Baf deficiency by performing combinational indexing-based single-cell RNA sequencing (scRNA-seq) of all brain cells (Fig. 1a). To this end, we collected third-instar larval brains from control larvae and larvae expressing Baf RNAi driven by elav-Gal4. Between two available Baf RNAi transgenes (BL36108 and KK102013), we used only the BL36108 line for scRNA-seq due to the off-target identified in the KK102013 line. Additionally, we confirmed the efficiency of the Baf RNAi (BL36108) transgene (Supplementary Fig. 1a, b). Thus, hereafter, “Baf RNAi” refers to the BL36108 transgenic line unless otherwise specified. From two pooled replicates for each condition, a total of 6365 cells (3176 control larval brain cells and 3189 Baf KD larval brain cells) passed our quality control criteria (Supplementary Fig. 1c–e; Supplementary Table 2). We applied a graph-based unsupervised clustering algorithm to systematically characterize the distinct cell types in the larval brain, resulting in 14 distinct clusters that were organized into 7 cell types (Supplementary Fig. 1c). After excluding glial cells, we subclustered 5949 neuronal cells into 13 distinct clusters that were organized into 5 cell types based on the expression of known cell type-specific markers (Fig. 1b, c and Supplementary Fig. 1f). Since all the samples were processed in the same batch, our scRNA-seq data showed minimal batch effects (Supplementary Fig. 1g–i). The two NB subsets (type I and II NBs) expressing pan-NB markers (dpn and N)47,48 were distinguished by the expression of ase (type I NB)49 and pnt (type II NB)50. The ganglion mother cell (GMC) population was defined by the expression of a canonical GMC marker (pros)51,52 and the absence of NB marker expression. The intermediate neural progenitor (INP) markers (ham and erm)53 were highly expressed in the INP population. The neuronal population was identified by the expression of neuronal markers (nSyb, Syt1, and Snap25). Additional cell type-specific marker genes that were recently used to annotate cell types in previous scRNA-seq studies54,55 were also considered in the grouping of cells into 5 different types (Fig. 1c).
a A schematic diagram of the combinatorial indexing-based scRNA-seq pipeline for third-instar larval brains. b UMAP plots showing all cells individually colored according to their cell type (top) or treatment condition (bottom). c Heatmap displaying the Z scores of the mean normalized expression of cell type-specific markers in each cell type. d Bar plot showing the AUC of each cell type indicating the separability of the control and Baf KD cells according to their transcriptomes. The number of each cell type and DEGs are shown in the additional table (right). e GO enrichment analysis of upregulated (orange) and downregulated (green) genes in type I NBs lacking Baf. f Heatmap showing the Z scores of the mean normalized expression of neurite-related genes that were upregulated in type I NBs lacking Baf. g Representative images of cultured primary type I NBs that were isolated from control larval brains (top, ase-Gal4/+;UAS-mCD8GFP,UAS-H2A.mRFP/+) or from larval brains expressing Baf RNAi (bottom, ase-Gal4/+;UAS-mCD8GFP,UAS-H2A.mRFP/UAS-Baf RNAi). Cultured primary type I NBs were visualized by the expression of the fluorescent plasma membrane marker protein, mCD8GFP. The left three panels show serial z-stack images, and the right panels show images processed with maximum intensity projection (M.I.P.) (Scale bar, 5 μm).
To determine which cell types in larval brains were most affected by Baf deficiency, we first quantified the transcriptomic differences in each cell type between the two conditions with respect to the number of differentially expressed genes (DEGs) and the separability of Baf KD and control cells according to their transcriptomes. Based on these two measures, type I NBs exhibited the most prominent changes in response to Baf deficiency (Fig. 1d and Supplementary Fig. 1j). Functional enrichment analysis of the DEGs in type I NBs revealed enrichment of biological processes that are crucial for the functionality and morphogenesis of differentiated neurons among the upregulated genes in the Baf KD group and enrichment of biological processes that are essential for cellular division among the downregulated genes in the Baf KD group (Fig. 1e). These data suggest that Baf KD specifically induces an aberrant shift in the transcriptional profiles of type I NBs toward those of differentiated neurons, thereby resulting in impaired formation of neural progenitor cell lineages. Notably, the biological processes that were enriched in the upregulated genes in the Baf KD group included genes that are involved in the regulation of neuronal morphology, such as ImpL2, RhoGAP93B, Snap25, for, CadN2, LanA, and scro (Fig. 1f). This finding is consistent with the observations that, compared with cultured control type I NBs, cultured primary type I NBs isolated from third-instar larval brains expressing Baf RNAi often exhibited bulging of the plasma membrane, failure to maintain a circular morphology, and increased diameter (Fig. 1g and Supplementary Fig. 1k, l). Taken together, these results demonstrate that Baf KD induces prominent transcriptional changes, particularly in type I NBs.
Baf KD in type I NBs causes defective anchoring of heterochromatin to the INM and impairs the formation of intact NBs and their neural progenies
We next explored the cellular defects of type I NBs lacking Baf. We first examined whether the subnuclear distribution of heterochromatin is altered by Baf deficiency through immunostaining for H3K9me3, a representative marker of constitutive heterochromatin56, in cultured primary type I NBs. We observed that heterochromatin labeled by H3K9me3 in control type I NBs tended to localize near the nuclear periphery, whereas that in type I NBs lacking Baf was either abnormally localized and enriched in the center of the nucleus (data not shown) or discontinuously scattered throughout the entire genome (Fig. 2a), indicative of defective subnuclear positioning of chromatin in Baf-deficient type I NBs.
a Representative images of immunostaining for H3K9me3 with Hoechst staining in cultured primary control type I NBs and type I NBs expressing Baf RNAi that were isolated from the brains of third-instar larvae (ase-Gal4/+;UAS-mCD8GFP/+ and ase-Gal4/+;UAS-mCD8GFP/UAS-Baf RNAi). NBs are visualized by the expression of the fluorescent plasma membrane marker protein mCD8GFP. The white dashed lines in the panels indicate the outlines of the inner nuclear membrane (Scale bar, 10 μm). b Representative images of control type I NBs (top, ase-Gal4/+;UAS-mCD8GFP/+) or type I NBs expressing Baf RNAi (bottom, ase-Gal4/+;UAS-mCD8GFP/UAS-Baf RNAi) in the brains of third-instar larvae. NBs are visualized by the expression of the fluorescent plasma membrane marker protein mCD8GFP. The white dashed lines in the left panels indicate the outlines of the brain lobes. Magnified images of type I NBs indicated by red arrowheads in the left panel are presented in the right panels (Scale bars, 20 μm). c Quantification of the number of type I NBs located in the apical cortex of third-instar larval brain lobes expressing the transgenes indicated in (b). **P < 1.0 × 10−2 according to Student’s t-test; error bars, mean ± SEM; the number of brain lobes tested was as follows: Control = 3 brain lobes, Baf KD = 3 brain lobes. d Quantification of the number of neural progenies derived from type I NBs located in the apical cortex of third-instar larval brain lobes expressing the transgenes indicated in (b). ****P < 1.0 × 10−4 according to Student’s t-test; error bars, mean ± SEM; the number of brain lobes tested was as follows: Control = 10 NBs, Baf KD = 6 NBs. e Representative images of BrdU (magenta) staining in control type I NBs and type I NBs expressing Baf RNAi in the brains of third-instar larvae. (ase-Gal4/+;UAS-mCD8GFP/+ and ase-Gal4/+;UAS-mCD8GFP/UAS-Baf RNAi). Magnified images of the red squares in the leftmost panels are presented in the second left panels. The white dashed lines in the panels indicate the outlines of the NBs (Scale bars, 20 μm). f Quantification of the proportion of BrdU-positive type I NBs to total type I NBs located in the apical cortex of third-instar larval brain lobes expressing the transgenes indicated in (e). ****P < 1.0 × 10−4 according to Student’s t-test; error bars, mean ± SEM; the number of brains tested is as follows: Control = 10 brain lobes, Baf KD = 7 brain lobes.
Then, we investigated the phenotypic changes of type I NBs in third-instar larval brains (Supplementary Fig. 2a) due to Baf deficiency. To this end, we knocked down Baf by ase-Gal449-driven expression of the Baf RNAi transgene in type I NBs of Drosophila. The number of type I NBs and their neural progenies were significantly decreased in brains expressing Baf RNAi compared to control brains (Fig. 2b–d), which was similarly observed in brains expressing the other Baf RNAi (KK102013), having one off-target (data not shown). Importantly, more than half of the NBs expressing either Baf RNAi showed marked defects in the formation of NBs and their neural progenies (magnified images in Fig. 2b and data not shown), indicative of defects in the self-renewal of type I NBs and the subsequent formation of neural progenitor cells. On the other hand, since our scRNA-seq data showed that Baf KD induced prominent transcriptional changes, particularly in type I NBs, we examined whether Baf KD induces cellular defects specifically in type I NBs but not in type II NBs. For this purpose, we selectively knocked down Baf in type II NBs using wor-Gal4 together with ase-Gal80 to prevent Gal4 expression in type I NBs57. Baf KD in type II NBs had a very marginal but not statistically significant effect on the formation of type II NBs and their neural progenies (Supplementary Fig. 2b–d), indicating that type I NBs are particularly vulnerable to the reduced dosage of the Baf gene during development of the Drosophila nervous system.
According to previous studies, Baf deficiency leads to defects in general cell division in both cultured mammalian cells and Drosophila26,58,59, which is consistent with our findings of defective self-renewal in type I NBs lacking Baf (Fig. 2b, c). Consistent with these findings, live imaging of cultured primary type I NBs expressing Baf RNAi displayed defective cell division accompanied by ectopically increased histone H2A levels compared to those of cultured control type I NBs (Supplementary Fig. 2e). In addition to the elevated levels of histone H2A, we also observed increased DNA contents in type I NBs lacking Baf compared to those in the control group. (Supplementary Fig. 2f, g). Increased DNA contents are indicative of defective cell division resulting from defective chromosomal segregation during cell division60. Furthermore, the significant reduction in the number of cells that contained phosphorylated histone, a specific marker of both mitosis and meiosis61, among type I NBs in larval brains expressing Baf KD also supported the occurrence of defective cell division in type I NBs lacking Baf (Supplementary Fig. 2h, i). We thus inferred that the increased levels of histone H2A are accompanied by an increase in DNA content in type I NBs deficient for Baf to maintain chromosomal structure within the nucleus. To further support this, we examined another histone protein, histone H3, and found that the level of histone H3 was also increased in type I NBs lacking Baf (Supplementary Fig. 2j). Overall, we concluded that heightened levels of histone H2A could be considered a cellular feature of defective cell division, consistent with the increased DNA contents in type I NBs lacking Baf. To further confirm the defect in the division of type I NBs caused by Baf deficiency, we explored whether the proliferation of NBs is indeed impaired by Baf deficiency by performing BrdU labeling of NBs in larval brains lacking Baf and those of controls. The proportion of BrdU-positive type I NBs among the total type I NBs in larval brains expressing Baf RNAi was significantly decreased compared to the control (Fig. 2e, f), indicating the defective proliferation of type I NBs caused by Baf deficiency. Taken together, Baf KD in type I NBs causing defective anchoring of heterochromatin to the INM impairs the formation of intact NBs and their neural progenies along with defective cell division.
Type I NB-specific neurodevelopment is substantially impaired by Baf KD, leading to a marked reduction in excitatory neurons in the adult stage
We next examined the developmental trajectories of type I and II NBs during neurogenesis in the brains of control larvae and larvae expressing Baf RNAi by using partition-based graph abstraction (PAGA)62. In the type I and II NB lineages, PAGA predicted a linear trajectory (type I NB – GMC – neuron or type II NB – INP – GMC – neuron) under both conditions (Fig. 3a). This prediction is consistent with the established temporal fate specification of NBs63. We confirmed the robustness of our predicted trajectories using Palantir45 (Supplementary Fig. 3a) and further analyzed the Baf KD-induced changes in expression dynamics along these developmental trajectories by aligning the two trajectories under both conditions. Similar to the analysis of regulator expression dynamics in the control group, the two trajectories of type II NBs were largely well aligned (Fig. 3b). However, we identified an off-diagonal alignment in early pseudotime for the trajectories of type I NBs (Fig. 3b). We then evaluated whether the perturbed linear trajectory of type I NBs compared to that of type II NBs is attributed to changes in the expression of related genes affected by Baf deficiency. To this end, we sorted the genes into 5 modules and conducted Gene Ontology (GO) analysis (Supplementary Fig. 3b, c). Within the linear trajectory of type I NBs, Baf deficiency disrupted the expression of a subset of transcriptional regulators that are associated with biological processes, including neuroblast fate determination (Dr64), neural precursor cell proliferation (SoxN65), regulation of cell cycle (cycA66, E2f167), chromatin remodeling (HmgD68), and chromatin assembly or disassembly (polybromo69) in module M1 (Supplementary Fig. 3d). Furthermore, genes involved in the negative regulation of cell fate specification (aop50,70, med471, and med1071) and negative regulation of cell proliferation (hfp70) were upregulated in the type I NB linear trajectory (module M2) (Supplementary Fig. 3d). Conversely, along the linear trajectory of type II NBs, Baf deficiency did not induce noticeable changes in the expression of the genes identified in modules M1 and M2 of the linear trajectory of type I NBs (Supplementary Fig. 3d). Taken together, these data indicate that Baf deficiency specifically impairs the fate specification of cells derived from type I NBs during neurodevelopment.
a PAGA graphs showing connectivity between cell types. The values on the right of each edge indicate the connectivity between cell types. b Dissimilarity matrices of global alignments of developmental trajectories of type I (top) and II (bottom) NB lineages. The histograms on each axis illustrate the individual cell types. c Representative images of immunostaining for Dpn (teal) and Pros (magenta) in control type I NBs and type I NBs expressing Baf RNAi in the brains of third-instar larvae (ase-Gal4/+;UAS-mCD8GFP/+ and ase-Gal4/+;UAS-mCD8GFP/UAS-Baf RNAi). The white dashed lines indicate the outlines of NBs (first column) and neural progenies (second column) (Scale bar, 20 μm). d Representative images of immunostaining for Dpn (teal) and elav (magenta) in type I NBs in the brains of third-instar larvae expressing the transgenes indicated in (d). The white dashed lines indicate the outlines of NBs (first column) and neural progeny cells (second column) (Scale bar, 20 μm). e Representative images of immunostaining for TH in adult brains of the following genotypes: UAS-Baf RNAi/+, elav-Gal4/UAS-Baf RNAi, ase-Gal4/+;UAS-Baf RNAi/+, and wor-Gal4,ase-Gal80/+;UAS-Baf RNAi/+. The red arrowheads indicate dopaminergic neurons (Scale bar, 40 μm). f Quantification of the number of dopaminergic neurons in the brains of flies expressing the indicated transgenes (UAS-Baf RNAi/+, elav-Gal4/+;UAS-Baf RNAi, ase-Gal4/+;UAS-Baf RNAi/+, and wor-Gal4,ase-Gal80/+;UAS-Baf RNAi/+). N.S., not significant; ****P < 1.0 × 10−4 by one-way ANOVA with Tukey’s post hoc test; error bars, mean ± SEM; UAS-Baf RNAi/+ = 7 brains, elav-Gal4 > Baf RNAi = 3 brains, type I-Gal4 > Baf RNAi = 5 brains, and type II-Gal4 > Baf RNAi = 5 brains. g Representative images showing the distribution and pattern of glutamatergic neurons in adult brains harboring the following genotypes: OK371-QF2/+;QUAS-, mCD8GFP/UAS-Baf RNAi, OK371-QF2/elav-Gal4;QUAS-mCD8GFP/UAS-Baf RNAi, OK371-QF2/ase-Gal4;QUAS-mCD8GFP/UAS-Baf RNAi, and OK371-QF2/wor-Gal4,ase-Gal80;QUAS-mCD8GFP/UAS-Baf RNAi. (Scale bar, 50 μm). h Quantification of the number of glutaminergic neurons in the brains of flies expressing the indicated transgenes (OK371-QF2/+;QUAS-mCD8GFP/UAS-Baf RNAi, OK371-QF2/elav-Gal4;QUAS-mCD8GFP/UAS-Baf RNAi, OK371-QF2/ase-Gal4;QUAS-mCD8GFP/UAS-Baf RNAi, and OK371-QF2/wor-Gal4,ase-Gal80;QUAS-mCD8GFP/UAS-Baf RNAi). N.S., not significant; **P < 1.0 × 10−2 by one-way ANOVA with Tukey’s post hoc test; error bars, mean ± SEM; n = 6 brains.
To experimentally validate the impaired fate specification of neural progenies derived from type I NBs caused by Baf deficiency, we first examined whether the formation of the polarity axis of NBs, which is important for their asymmetric division72,73,74 and the production of their neural progenies, is affected by Baf deficiency in type I NBs. According to previous studies, the asymmetric distribution of proteins that regulate self-renewal or differentiation of daughter cells is essential for the asymmetric cell division of NBs75,76. To achieve this, atypical protein kinase C (aPKC), a component of the par complex, localizes to the apical cortical region and restricts basal domain components, such as Miranda (Mira), to the basal region of NBs72,73. Mira, an adaptor protein of Pros and Brat, localizes to the basal cortex through direct aPKC phosphorylation, which contributes to the polarized distribution of fate determinants during asymmetric cell division of NBs73,77. Based on these findings, we examined the distribution and expression of aPKC and Mira in cultured primary type I NBs lacking Baf by immunostaining. While asymmetric and polarized distributions of both aPKC (apical distribution) and Mira (basal distribution) were detected in control Type I NBs, aPKC and Mira were almost undetectable in type I NBs deficient for Baf (Supplementary Fig. 3e). We could occasionally observe detectible signals of Mira in type I NBs lacking Baf, and in these cells, Mira exhibited a dispersed and unpolarized distribution (data not shown) instead of the asymmetric and polarized distribution that was observed in the control. The perturbed formation of the polarity axis in NBs suggested that asymmetric cell division in type I NBs is impaired by Baf deficiency, which contributes to the malformation of neural progenitor cell lineages. We then examined whether Baf deficiency in type I NBs leads to alterations in the expression of cell fate determinants in individual neural progenies derived from type I NBs. For this purpose, we labeled type I NBs and their neural progenies by immunostaining Dpn and cell fate determinants (Pros for GMC and elav for neuron), respectively, in control larval brains and larval brains lacking Baf. Notably, the neural progenies derived from type I NBs lacking Baf showed decreased level of each cell fate determinant compared to that of the control (Fig. 3c, d). These results suggest that Baf deficiency impairs the fate specification of neural progeny derived from type I NBs.
We then examined whether Baf deficiency in type I NBs during larval development leads to alterations in the cellular composition of the nervous system in adult flies. To this end, we first labeled representative excitatory neurons, such as dopaminergic (DA) and glutamatergic neurons, in control adult brains and adult brains expressing Baf RNAi and assessed their numbers and distribution patterns. Immunostaining with an anti-tyrosine hydroxylase (TH) antibody revealed a significant reduction in the number of DA neurons in response to both pan-neuronal and type I NB-specific KD of Baf compared to that in the control group (Fig. 3e, f). On the other hand, type II NB-specific KD of Baf had no effect on either the number or distribution pattern of DA neurons (Fig. 3e, f). We also used the Q-system (OK371-QF2/+;QUAS-mCD8GFP/+) to label glutamatergic neurons in fly brains expressing Baf RNAi. Similar to DA neurons, the number of glutamatergic neurons in type I NBs was significantly decreased in fly brains pan-neuronally expressing Baf RNAi and those expressing Baf RNAi in type I NBs, but not in those expressing Baf RNAi in type II NBs (Fig. 3g, h). We then examined the number of other types of neurons, such as GABAergic, cholinergic, and serotonergic neurons, in adult brains pan-neuronally expressing Baf RNAi by immunostaining. Due to the high population density of GABAergic and cholinergic neurons in the central brain, we specifically counted the numbers of these neurons that were located near brain regions, such as the AMMC and SEZ, primarily associated with the subset of neurons comprising the neural circuit, which is responsible for grooming behavior78,79. Consequently, we assessed the numbers of these neurons located in the aforementioned regions of the Baf KD adult Drosophila brains and compared them to those in the control group. However, there were no significant changes in the numbers of GABAergic, cholinergic, or serotonergic neurons between the Baf KD and control brains (Supplementary Fig. 3f, g). Taken together, these data demonstrate that Baf KD impairs the formation of type I NB lineages and causes a substantial reduction in the number of excitatory neurons, such as DA and glutamatergic neurons, in adult brains.
Baf KD in type I NBs induces distinct behavioral abnormalities in adults
Next, we explored the functional consequences of the aforementioned defects in the NB lineage formation caused by Baf KD. To this end, we characterized the behavioral phenotypes of adult flies expressing Baf RNAi driven by elav-Gal4. First, we observed that the majority (75%) of flies expressing Baf RNAi exhibited abnormal wing postures (droopy or held-up wing) (Supplementary Fig. 4a). The flies expressing Baf RNAi also displayed two additional behavioral abnormalities, characterized as excessive grooming and freezing-like behavior (termed “immobility behavior”) (Fig. 4a). To better characterize these unique behavioral abnormalities, we placed an individual male fly in a circular arena and monitored its movement for 10 min. The Baf-deficient flies exhibited a mixture of these two behavioral abnormalities. We considered a fly to be exhibiting a particular abnormal behavior when that behavior was observed for greater than one-third of the total recording time. In severe cases, flies exhibited both behavioral abnormalities, and these flies were labeled “mixed”. Notably, the majority of the flies lacking Baf exhibited aberrant behaviors (Fig. 4b, c). In line with these findings, adult flies pan-neuronally expressing Baf RNAi exhibited significantly increased total time spent on grooming and immobility compared to the control flies (Fig. 4d). Largely due to these unique behavioral abnormalities, flies expressing Baf RNAi barely walked and thus showed a reduced mean velocity (Supplementary Fig. 4b–d).
a Representative ethograms showing the distinct behaviors of control flies (elav-Gal4/+) and Baf KD flies (elav-Gal4/UAS-Baf RNAi). Different colors are used to distinguish different aspects of fly behavior. n = 5. b The proportion of control flies (elav-Gal4/+) or Baf KD flies (elav-Gal4/UAS-Baf RNAi) exhibiting aberrant behavior. The number of flies tested was as follows: Control = 5 flies and Baf KD = 5 flies. c Stacked bar graph showing the proportions of distinct behavioral abnormalities in control flies (elav-Gal4/+) or Baf KD flies (elav-Gal4/UAS-Baf RNAi). The number of flies tested was as follows: Control = 5 flies and Baf KD = 5 flies. d Box-and-whisker plots representing the 10th–90th percentiles with the mean value (horizontal line) of total time spent on grooming and immobility (elav-Gal4/+ or elav-Gal4/UAS-Baf RNAi) for 10 min. ****P < 1.0 × 10−4 by Student’s t-test; n = 5 flies. e Representative ethograms showing distinct behaviors of flies expressing the indicated genotypes (UAS-Baf RNAi/+, ase-Gal4/+;UAS-Baf RNAi/+, and wor-Gal4,ase-Gal80/+;UAS-Baf RNAi/+). Different colors are used to distinguish different aspects of fly behavior. n = 5. f Box-and-whisker plots representing the 10th–90th percentiles with the mean value (horizontal line) of total time spent on grooming and immobility in flies expressing the transgenes indicated in (e) for 10 min. N.S., not significant; ****P < 1.0 × 10−4 by one-way ANOVA with Tukey’s post hoc test; n = 5 flies.
We next examined whether the observed behavioral abnormalities in Baf-deficient flies are primarily attributed to the neurodevelopmental defects that were characterized above prior to the maturation of the nervous system in the adult stage. To this end, we conditionally knocked down Baf before or after the eclosion of adult flies using Gal80TS, which is a temperature-sensitive negative regulator of Gal4 [temporal and regional gene expression targeting (TARGET) system]80, and assessed the aberrant behaviors of the adult flies. Conditional Baf KD before the eclosion of adult flies induced significant increases in the total time spent on grooming and immobility, whereas conditional Baf KD during the adult stage did not induce significant changes in grooming or immobility (Supplementary Fig. 4e). Consistently, we found that Baf KD in differentiated mature neurons or glial cells driven by nSyb- and repo-Gal4, respectively, did not induce behavioral abnormalities (Supplementary Fig. 4f, g). We further examined whether Baf KD in type I NBs recapitulates the unique behavioral abnormalities caused by the elav-Gal4-driven KD of Baf. We found that Baf KD in type I NBs, but not in type II NBs, markedly increased the behavioral abnormalities (Fig. 4e, f). Collectively, these data indicate that Baf deficiency in type I NBs during neurodevelopment prior to nervous system maturation leads to distinct behavioral abnormalities in the adult stage.
Transcriptional downregulation of tsh, which interacts with beta-catenin, caused by Baf deficiency in type I NBs is associated with behavioral abnormalities in adult flies
We next investigated the molecular mechanism by which Baf KD-mediated transcriptional changes in type I NBs during neurodevelopment contribute to acquired behavioral abnormalities. To this end, we performed a series of genetic screenings that involved knocking down and overexpressing candidate genes that were identified as DEGs in type I NBs in our scRNA-seq data. Among the 275 DEGs (Supplementary Table 3), there were available transgenic fly lines that corresponded to 117 of the genes (Fig. 5a). As an initial screen, we individually overexpressed 30 upregulated genes and knocked down 87 downregulated genes in type I NBs and examined whether these flies exhibited aberrant wing postures similar to those of Baf KD flies (Supplementary Fig. 4a). Our initial screening revealed that aberrant wing postures were induced by knocking down 19 of the downregulated genes (teashirt [tsh], Spindly, bellwether [blw], moleskin [msk], CG5033, Chromatin assembly factor 1 p55 subunit [Caf1-55], ventral veins lacking [vvl], microtubule star [mts], Ubiquitin-like activating enzyme 2 [Uba2], U4-U6 small nuclear riboprotein factor 60K [U4-U6-60K], mars, fascetto [feo], Heat-shock-protein-70Bc [Hsp70Bc], Rap1 GTPase [Rap1], puffyeye [puf], Zinc transporter 49B [Znt49B], Foxo, Ranbp9 and CG43340) or by overexpressing 2 of the upregulated genes (Forkhead box K [FoxK] and Ods-site homeobox [OdsH]) in type I NBs (red and turquoise dots in Fig. 5b). We further evaluated whether downregulated 19 genes inducing wing posture problems in adult flies were correlated with heterochromatin remodeling/anchoring by conducting GO analysis of these genes compared to the other genes in the group of 117 genes. We found that among the GO terms specifically annotated to the 19 genes, those related to chromosome organization, such as chromosome segregation and nucleosome mobilization, were particularly enriched (Supplementary Fig. 5a). This enrichment suggested that the reduced gene expression may contribute to defective heterochromatin anchoring in type I NBs lacking Baf (Supplementary Fig. 5a).
a The overall workflow for identifying downstream mediators of Baf-dependent regulation of type I NB-specific neurodevelopment. b Scatter dot plot showing the proportion of flies exhibiting aberrant wing postures induced by type I NB-specific expression of up- or downregulated DEGs that were identified in type I NBs. n ≥ 50 flies. Every scattered dot in the graph indicates KD of an indicated gene in type I NBs. Among these dots, red dots, but not turquoise dots, showed behavioral abnormalities in the adult stage (see Figure S5b for details). c Representative ethograms showing distinct behaviors of adult flies expressing transgenes as follows (ase-Gal4/+;UAS-Baf RNAi/+, ase-Gal4/UAS-tsh;UAS-Baf RNAi/+, and ase-Gal4/UAS-msk;UAS-Baf RNAi/+). Different colors are used to distinguish different aspects of fly behavior. n = 5. d Box-and-whisker plots representing the 10th–90th percentiles with the mean value (horizontal line) of total time spent on grooming and immobility in flies expressing the transgenes indicated in (c) for 10 min. N.S., not significant; ****P < 1.0 × 10−4 by one-way ANOVA with Tukey’s post hoc test; n = 5 flies. e Representative ethograms showing distinct behaviors of adult flies expressing transgenes as follows (ase-Gal4/+;UAS-Baf RNAi/+, ase-Gal4/UAS-Arm;UAS-Baf RNAi/+, UAS-Arm.S10/+;ase-Gal4/+;UAS-Baf RNAi/+, and ase-Gal4/+;UAS-Baf RNAi/UAS-Arm RNAi). Different colors are used to distinguish different aspects of fly behavior. n = 5. f Box-and-whisker plots representing the 10th–90th percentiles with the mean value (horizontal line) of total time spent on grooming and immobility in flies expressing the transgenes indicated in (e) for 10 min. N.S., not significant; ****P < 1.0 × 10−4 by one-way ANOVA with Tukey’s post hoc test; n = 5 flies.
We subsequently performed successive screening to examine whether genetic modulation of these 21 genes can induce behavioral abnormalities in the adult stage. Among the 21 genes, KD of 11 downregulated genes (tsh, Spindly, blw, msk, CG5033, Caf1-55, vvl, mts, Uba2, U4-U6-60K, and mars) in type I NBs induced aberrant behaviors in the adult flies (Fig. 5b (red dots) and Supplementary Fig. 5b) compared to the controls. Notably, increased behavioral abnormalities comparable to those induced by Baf KD were observed only in flies expressing tsh RNAi or Spindly RNAi (Supplementary Fig. 5c). We further examined whether genetic modulation of these 11 genes can suppress Baf KD-induced behavioral abnormalities. Among the 11 downregulated genes, only 4 of these genes had available overexpression transgenic fly lines. Overexpression of tsh or msk significantly suppressed Baf KD-induced behavioral abnormalities, whereas overexpression of Caf1-55 or mts did not induce noticeable changes in the total time spent on grooming and immobility (Fig. 5c, d and Supplementary Fig. 5d, e), suggesting that Baf-dependent transcriptional control of tsh and msk is crucial for type I NB-specific neurodevelopment. We concluded that, between tsh and msk, tsh is a more potent factor in mediating Baf-dependent control of NB lineage development since KD of tsh, but not KD of msk, induced behavioral abnormalities comparable to those induced by KD of Baf (Supplementary Fig. 5b and see “Discussion” for details).
We next explored additional molecules/signaling pathways that could contribute to the Baf-dependent control of NB lineage development by interacting with tsh and/or msk. To this end, we examined the protein‒protein interaction (PPI) networks of tsh and msk using the Search Tool in the Retrieval of Interacting Genes/Proteins (STRING) web database and identified Armadillo [Arm], a Drosophila homolog of beta-catenin in the Wnt signaling pathway, as a factor that interacts with both tsh and msk. Previous studies reported that the zinc finger TF tsh binds to the C-terminus of Arm and is associated with trunk-specific modulation of the Wnt/beta-catenin pathway in Drosophila embryos81,82 and that msk, which is a Drosophila homolog of importin-7, induces the nuclear transport of Arm to regulate the Wnt signaling pathway83.
Given the well-characterized functions of the Wnt/beta-catenin pathway in the regulation of neural cell fate determination84,85, we speculated that Arm could contribute to Baf-dependent neurodevelopment through its interaction with tsh and/or msk. To experimentally validate this speculation, we first evaluated whether genetic disruption of Arm in type I NBs induces behavioral abnormalities in adult flies similar to those induced by Baf KD. Compared with Baf KD, Arm KD increased the total time spent on grooming and immobility (Fig. 4e, f and Supplementary Fig. 5f, g), which was consistent with our speculation. We then examined whether overexpression of Arm can ameliorate Baf KD-induced behavioral abnormalities. To this end, we individually overexpressed Arm or Arm.S10, a constitutively active form of Arm86, or knocked down Arm in type I NBs expressing Baf RNAi, and evaluated behavioral patterns in the adult stage. Notably, overexpressing Arm or Arm.S10 significantly decreased the total time of Baf KD-induced behavioral abnormalities (Fig. 5e, f), which further validates our speculation. On the other hand, KD of Arm did not cause any noticeable changes in Baf KD-induced behavioral abnormalities (Fig. 5e, f), suggesting that Baf KD-induced behavioral abnormalities are already too severe to be further impaired.
Transcriptional downregulation of tsh, which interacts with beta-catenin, caused by Baf deficiency in type I NBs is responsible for the type I NB lineage development
We next examined whether KD of tsh or Arm recapitulates Baf KD-induced defects in the development of type I NB lineages. However, KD of tsh or Arm in type I NBs barely impaired the formation of type I NBs and their neural progenies (Supplementary Fig. 6a–c), indicating that KD of tsh or Arm alone is not sufficient to impair the symmetric/asymmetric division of type I NBs. Similarly, the expression of msk RNAi in type I NBs did not show any noticeable changes in the number of type I NBs and the formation of their neural progenies (Supplementary Fig. 6d, e). To further characterize the cellular defects caused by KD of tsh or Arm in type I NBs, we next examined whether KD of tsh or Arm in type I NBs leads to dysregulation of cell fate determinants in the neural progenies of type I NBs. Notably, the neural progenies derived from type I NBs lacking tsh or Arm showed reduced level of Pros, compared to those of control group (Fig. 6a), similar to what was observed in the case of Baf KD. These results collectively suggest that genetic disruption of tsh or Arm in type I NBs induces detrimental cellular changes in type I NBs and their neural progenies as well as dysregulation of cell fate determinants in the neural progenies, although these detrimental cellular changes are insufficient to disrupt the symmetric/asymmetric division of type I NBs.
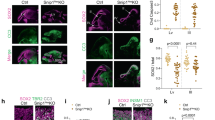
a Representative images of immunostaining for Dpn (teal) and Pros (magenta) in control type I NBs and type I NBs expressing Baf RNAi (Baf KD), tsh RNAi (tsh KD), and Arm RNAi (Arm KD) in brains of third-instar larvae (ase-Gal4/+; UAS-mCD8GFP/+, ase-Gal4/+;UAS-Baf RNAi/UAS-mCD8GFP, ase-Gal4/+;UAS-tsh RNAi/UAS-mCD8GFP, and ase-Gal4/+;UAS-Arm RNAi/UAS-mCD8GFP). The white dashed lines indicate the outlines of NBs (second row) and neural progenies (third row) (Scale bar, 20 μm). b Representative images of immunostaining for H3K9me3 (magenta) with Hoechst staining in cultured primary control type I NBs and type I NBs expressing Baf RNAi (Baf KD), tsh RNAi (tsh KD), and Arm RNAi (Arm KD) that were isolated from the brains of third-instar larvae (ase-Gal4/+; UAS-mCD8GFP/+, ase-Gal4/+;UAS-Baf RNAi/UAS-mCD8GFP, ase-Gal4/+;UAS-tsh RNAi/UAS-mCD8GFP, and ase-Gal4/+;UAS-Arm RNAi/UAS-mCD8GFP). NBs were visualized by expression of the fluorescent plasma membrane marker protein mCD8GFP. The white dashed lines in the panels show the outlines of the inner nuclear membrane (Scale bar, 10 μm). c Stacked bar graph showing the proportion of mean intensity of heterochromatin localized in three zones of cultured primary type I NBs expressing the transgenes indicated in (b). The number of type I NBs tested was as follows: Control = 6, Baf KD = 7, tsh KD = 11, and Arm KD = 5. d Scatter dot plots displaying the proportion of mean intensity of heterochromatin localized in the peripheral zone of type I NBs expressing the transgenes indicated in (b). ***P < 1.0 × 10−3; ****P < 1.0 × 10−4 by one-way ANOVA with Tukey’s post hoc test; error bars, mean ± SEM. The number of type I NBs tested was as follows: Control = 6, Baf KD = 7, tsh KD = 11, and Arm KD = 5. e Scatter dot plots displaying the proportion of mean intensity of heterochromatin localized in the central zone of type I NBs expressing the transgenes indicated in (b). **P < 1.0 × 10−2; ***P < 1.0 × 10−3 by one-way ANOVA with Tukey’s post hoc test; error bars, mean ± SEM. The number of type I NBs tested was as follows: Control = 6, Baf KD = 7, tsh KD = 11, and Arm KD = 5. f Representative images of immunostaining for Baf (magenta) and tsh (teal) in cultured primary type I NBs that were isolated from the brains of third-instar larvae (ase-Gal4/+; UAS-mCD8GFP/+). NBs are visualized by expression of the fluorescent plasma membrane marker protein mCD8GFP. The white dashed lines in the panels show the outlines of the inner nuclear membrane (Scale bar, 10 μm).
We then wondered whether KD of tsh or Arm, which recapitulates Baf KD-induced defects in behavior and the formation of type I NB lineages also impairs the anchoring of heterochromatin to the INM. Quantification of the subnuclear positioning of heterochromatin (Supplementary Fig. 6f) was conducted as previously described87,88. Heterochromatin in type I NBs lacking tsh or Arm tended to localize to the center of the nucleus, comparable to that in type I NBs lacking Baf (Fig. 6b–e), suggesting that tsh and Arm are also needed to anchor heterochromatin to the INM, at least in type I NBs, rather than functioning downstream in the process of altered heterochromatin anchoring. To deepen our understanding of the molecular characteristics of key genes (Baf, tsh, and Arm) specifically in type I NBs, we examined the subcellular localization of these molecules in type I NBs by immunostaining. Notably, we detected Baf and tsh using their respective antibodies, revealing a pattern of colocalization in a region adjacent to DNA labeled by Hoechst staining within the nucleus of type I NBs (Fig. 6f). On the other hand, Arm was predominantly localized in the cytoplasmic region of type I NBs (Supplementary Fig. 6g), which is consistent with the previously described localization of Arm/beta-catenin in most cells in the absence of specific upstream cues89,90. We next examined whether there were any notable changes in the level or localization of key proteins between Arm, which is primarily localized in the cytoplasm, and the nuclear-localized Baf and tsh in type I NBs. However, the expression and localization of Arm in type I NBs did not noticeably change regardless of the presence of Baf or tsh (Supplementary Fig. 6h), and KD of Arm also did not induce alterations in the subcellular distribution of Baf and tsh in type I NBs compared to the control (Supplementary Fig. 6i). Overall, our immunohistochemistry data did not provide direct evidence of changes in the amount and/or localization of Arm and the nuclear-localized proteins Baf and tsh in type I NBs, despite the expected significant role of Arm in the formation of Type I NB lineages, as suggested by the genetic rescue data presented above (Fig. 5e, f).
To enhance our understanding of the mode-of-action of key genes engaged in anchoring heterochromatin to the INM in type I NBs, we examined whether KD of each key gene that colocalized to a region adjacent to heterochromatin in type I NBs affects the subcellular localization of the other key gene that colocalize to this region. In this regard, it should be noted that, in type I NBs lacking Baf showing altered subcellular localization of heterochromatin (Fig. 2a), immunostaining for tsh failed to detect signals for the endogenous tsh proteins (data not shown), consistent with our scRNA-seq data showing substantially decreased expression of tsh, but not Arm, in type I NBs (Supplementary Fig. 6j). Next, we investigated whether KD of tsh affects the subcellular localization of both Baf and heterochromatin in type I NBs. Notably, KD of tsh in type I NBs changed the subcellular localization of heterochromatin without affecting that of Baf (Supplementary Fig. 6k). Together with the pattern of tsh and Baf colocalization in the nucleus of type I NBs, these data collectively suggest that both Baf and tsh are required for anchoring heterochromatin to the INM in type I NBs, while the binding of heterochromatin to each of these proteins does not necessitate the presence of the other.
Finally, we wondered whether overexpression of tsh or Arm in type I NBs lacking Baf is capable of suppressing Baf KD-induced defects in the formation of type I NB lineages during neurodevelopment. Of note, overexpression of tsh or Arm markedly restored these numbers of type I NBs and their neural progenies impaired by Baf deficiency (Fig. 7a–c). Moreover, Baf deficiency altered the localization of heterochromatin in type I NBs, but overexpression of tsh or Arm restored the subnuclear position of heterochromatin to the peripheral region of the nucleus (Fig. 7d, e and Supplementary Fig. 7a, b). Furthermore, overexpression of tsh or Arm in type I NBs lacking Baf partially restored the expression level of Pros in the neural progenies derived from type I NBs (Supplementary Fig. 7c). Consistent with the finding that overexpression of tsh or Arm suppressed Baf KD-induced defects in the type I NB lineage during larval development, overexpression of tsh or Arm restored the number of DA neurons, which had been impaired by Baf deficiency (Supplementary Fig. 7d, e). Collectively, our genetic analyses identified tsh, which interacts with Arm, as a key downstream mediator of Baf in the development of type I NB cell lineages and as a crucial factor that complements the Baf-dependent control of heterochromatin anchoring.
a Representative images of control type I NBs and type I NBs expressing Baf RNAi (Baf KD), Baf RNAi + tsh (Baf KD + tsh OE), and Baf RNAi + Arm (Baf KD + Arm OE) in the brains of third-instar larvae (ase-Gal4/+;UAS-mCD8GFP/+, ase-Gal4/+;UAS-mCD8GFP/UAS-Baf RNAi, ase-Gal4/UAS-mCD4GFP;UAS-tsh/UAS-Baf RNAi, and ase-Gal4/UAS-Arm;UAS-mCD8GFP/UAS-Baf RNAi). Magnified images of the red squares in the upper panels are presented in the lower panels (Scale bars, 20 μm). b Quantification of the number of type I NBs located in the apical cortex of third-instar larval brain lobes expressing the transgenes indicated in (a). ***P < 1.0 × 10−3 according to one-way ANOVA with Tukey’s post hoc test; error bars, mean ± SEM; the number of brain lobes tested was as follows: Control = 3 brain lobes, Baf KD = 3 brain lobes, Baf KD + tsh OE = 5 brain lobes, and Baf KD + Arm OE = 5 brain lobes. c Quantification of the number of neural progenies derived from type I NBs located in the apical cortex of third-instar larval brain lobes expressing the transgenes indicated in (a). **P < 1.0 × 10−2, ****P < 1.0 × 10−4 by one-way ANOVA with Tukey’s post hoc test; error bars, mean ± SEM; the number of NBs tested was as follows: Control = 8 NBs, Baf KD = 4 NBs, Baf KD + tsh OE = 7 NBs, and Baf KD + Arm OE = 9 brain lobes. d Representative images of cultured primary type I NBs isolated from the brains of third-instar larvae expressing the transgenes indicated in (a). The white dashed lines in the panels show the outline of the inner nuclear membrane (Scale bar, 10 μm). e Stacked bar graph showing the proportion of mean intensity of heterochromatin localized in three zones of cultured primary type I NBs expressing the genes indicated in (a). The number of type I NBs tested was as follows: Control = 12, Baf KD = 10, Baf KD + tsh OE = 14, and Baf KD + Arm OE = 9.
Discussion
In this study, we demonstrated that Baf deficiency in type I NBs of Drosophila causing dysregulation of heterochromatin anchoring leads to impairment of type I NB-specific neurodevelopment and subsequent behavioral abnormalities. Notably, among the DEGs in type I NBs that were identified in our scRNA-seq data, our genetic analyses revealed that tsh, a zinc-finger TF that interacts with beta-catenin in the Wnt signaling pathway, is a key downstream mediator of Baf in the regulation of NB lineages and a complementing factor of Baf-dependent control of heterochromatin anchoring. We also found that tsh colocalizes with Baf in a region adjacent to heterochromatin in type I NBs, which may provide valuable clues for understanding the mode-of-action underlying the cooperative regulation of heterochromatin anchoring to the INM by Baf and tsh in the development of type I NB lineages.
Notably, Baf KD caused prominent changes in transcriptional profiles, particularly in type I NBs, although elav-Gal4 drives the expression of Baf RNAi globally throughout the central nervous system. This cell type-specific effect of Baf KD can be explained by the low Baf protein expression specifically in type I NBs. Although we think that the level of Baf expression in the central nervous system of Drosophila larvae may not be high, judging from the relatively small detection of Baf mRNA levels in our scRNA-seq data and the generally weak intensity of immunostaining with an anti-Baf antibody (data not shown), we excluded the possibility of low Baf expression specifically in type I NBs based on a previous study that used an anti-Baf antibody to show ubiquitous expression of endogenous Baf throughout entire brains of larvae32. Instead, it seems more likely that functionally redundant molecules of Baf (e.g., other chromatin-binding proteins), downstream mediators of Baf (e.g., tsh and Arm), and/or interactors of Baf (e.g., LEM domain-containing proteins) may be more abundant/active in other cell types, increasing the sensitivity of type I NBs to Baf KD. In this regard, considering our findings (Fig. 2a) and the findings of previous studies showing that Baf deficiency fails to generate intact progenies through mitotic cell division, we carefully speculate that cell type-specific effects may be critically regulated by the presence and/or amounts of tsh to the level comparable to or even more important than those of Baf, which is thought to be ubiquitously expressed and has essential mitotic functions.
Interestingly, certain processes involved in mammalian neurogenesis share similarities with those involved in Drosophila neurogenesis. In particular, the key principle of neurogenesis (e.g., asymmetric division accompanied by a polarized distribution of cell fate determinants) appears to be evolutionarily conserved. In the mammalian neocortex, various types of neural progenitors, such as radial glial cells, short neural precursors, and outer radial glial cells, can be found. These progenitors undergo asymmetric division, allowing them to both self-renew and produce neurons/intermediate neural progenitors91,92. Among them, radial glial cells in mammals largely undergo asymmetric cell division, similar to type I NBs in Drosophila93,94. As in Drosophila, the fate of neural progenitor cells in mammals is governed by the spatial and temporal TFs, enabling these cells to give rise to diverse types of neural progeny95,96. However, our understanding of the upstream regulators or regulatory processes governing the sequential expression of TFs during mammalian development remains relatively less understood. To bridge this knowledge gap, the mechanistic insights from our study may be broadly applicable beyond Drosophila and could be extended to the mammalian system. Notably, Banf1, a mammalian homolog of Baf, functions as a chromatin-anchoring protein located within the inner nuclear membrane26. In addition, a previous study demonstrated that deficiencies in Banf1 result in reduced expression of TFs that are associated with pluripotency in mouse embryonic stem cells and lead to defects in survival of both mouse and human embryonic stem cells97. Based on these findings, we speculated that Banf1 may also play an essential role in the development of neural progenitor cell lineages by mediating the subnuclear positioning of chromatin and regulating the transcription of genes that determine cell lineages. To experimentally validate this, scRNA-seq experiments and Banf1-deficient mouse studies should be conducted in a manner similar to our study. In addition to Banf1, a more focused study on tshz1, the mammalian counterpart of tsh, is needed to validate our findings in the mammalian context. Notably, previous research revealed that tshz1 functions as a transcriptional regulator of genes that are involved in neurodevelopment98,99. According to a previous study, tshz1 is involved in the differentiation and radial migration of neural progenitor cells within the olfactory bulb99. It will be interesting to see whether our findings are applicable to this process. We hope that the application of our findings to mammalian systems in this respect will provide a new direction for understanding the cell type-specific regulation of neurogenesis via the control of heterochromatin anchoring by specific molecular machineries, which primarily consist of Banf1 and tshz1.
Among the DEGs that were identified in type I NBs, our genetic analyses revealed that the downregulation of tsh or msk may be related to the Baf KD-induced phenotypes. Type I NB-specific overexpression of these genes significantly suppressed Baf KD-induced behavioral abnormalities (Fig. 5c, d). However, we focused on tsh in our study because KD of msk induced a distinct behavioral phenotype, tremor, that was not observed in adult flies lacking Baf or tsh (Supplementary Fig. 5b). The occurrence of distinct behavioral defects induced by msk KD suggested that msk KD involves additional molecular mechanisms other than those involving Arm. We suspect that the mitogen-activated protein kinase (MAPK) signaling pathway is a potential target based on previous reports indicating that msk, a Drosophila homolog of importin-7, is involved in the translocation of activated MAPK into the nucleus100,101. Given the well-characterized roles of MAPK signaling in the determination of neural cell fate102,103, dysregulation of MAPK signaling by msk KD in type I NBs may lead to abnormal neurodevelopment and distinct behavioral abnormalities. Since overexpression of msk markedly suppressed the Baf KD-induced phenotypes, it will be interesting to determine whether MAPK signaling also participates in the Baf-mediated regulation of the type I NB lineage development in addition to beta-catenin.
In summary, our study provides evidence that the heterochromatin anchoring-dependent transcriptional regulation of specific gene(s) may be essential for the formation of neural progenitor cell lineages and subsequent neurodevelopment. Moreover, our data provide new mechanistic insight that tsh interacting with Arm, of which expression is regulated by Baf in a cell type-specific manner, colocalizes with Baf in a region adjacent to heterochromatin in type I NBs and can complement the role of Baf in the formation of NB lineages through its overexpression, under conditions of Baf deficiency. Our unique approach of combining scRNA-seq and a series of genetic analyses will inspire other studies in exploring the regulatory mechanisms involved in the development of neural progenitor cell lineages.
References
Deneris, E. S. & Hobert, O. Maintenance of postmitotic neuronal cell identity. Nat. Neurosci. 17, 899–907 (2014).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Almeida, N. et al. Employing core regulatory circuits to define cell identity. EMBO J. 40, e106785 (2021).
Doe, C. Q. Temporal patterning in the Drosophila CNS. Annu. Rev. Cell Dev. Biol. 33, 219–240 (2017).
Holguera, I. & Desplan, C. Neuronal specification in space and time. Science 362, 176–180 (2018).
Grosskortenhaus, R., Robinson, K. J. & Doe, C. Q. Pdm and Castor specify late-born motor neuron identity in the NB7-1 lineage. Genes Dev. 20, 2618–2627 (2006).
Isshiki, T., Pearson, B., Holbrook, S. & Doe, C. Q. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell 106, 511–521 (2001).
Jafari, S. et al. Combinatorial activation and repression by seven transcription factors specify Drosophila odorant receptor expression. PLoS Biol. 10, e1001280 (2012).
Harr, J. C., Gonzalez-Sandoval, A. & Gasser, S. M. Histones and histone modifications in perinuclear chromatin anchoring: from yeast to man. EMBO Rep. 17, 139–155 (2016).
Zheng, H. & Xie, W. The role of 3D genome organization in development and cell differentiation. Nat. Rev. Mol. Cell Biol. 20, 535–550 (2019).
Kishi, Y. & Gotoh, Y. Regulation of chromatin structure during neural development. Front Neurosci. 12, 874 (2018).
Ye, Y. et al. Chromatin remodeling during in vivo neural stem cells differentiating to neurons in early Drosophila embryos. Cell Death Differ. 24, 409–420 (2017).
Chathoth, K. T. & Zabet, N. R. Chromatin architecture reorganization during neuronal cell differentiation in Drosophila genome. Genome Res. 29, 613–625 (2019).
Dixon, J. R. et al. Chromatin architecture reorganization during stem cell differentiation. Nature 518, 331–336 (2015).
Zhao, R., Bodnar, M. S. & Spector, D. L. Nuclear neighborhoods and gene expression. Curr. Opin. Genet Dev. 19, 172–179 (2009).
Guerreiro, I. & Kind, J. Spatial chromatin organization and gene regulation at the nuclear lamina. Curr. Opin. Genet Dev. 55, 19–25 (2019).
Montes de Oca, R., Andreassen, P. R. & Wilson, K. L. Barrier-to-Autointegration Factor influences specific histone modifications. Nucleus 2, 580–590 (2011).
Oh, H. S., Traktman, P. & Knipe, D. M. Barrier-to-autointegration factor 1 (BAF/BANF1) promotes association of the SETD1A histone methyltransferase with herpes simplex virus immediate-early gene promoters. mBio 6, e00345–00315 (2015).
Montes de Oca, R., Lee, K. K. & Wilson, K. L. Binding of barrier to autointegration factor (BAF) to histone H3 and selected linker histones including H1.1. J. Biol. Chem. 280, 42252–42262 (2005).
Dunlevy, K. L. et al. The PRR14 heterochromatin tether encodes modular domains that mediate and regulate nuclear lamina targeting. J. Cell Sci. https://doi.org/10.1242/jcs.240416 (2020).
Bian, Q., Anderson, E. C., Yang, Q. & Meyer, B. J. Histone H3K9 methylation promotes formation of genome compartments in Caenorhabditis elegans via chromosome compaction and perinuclear anchoring. Proc. Natl Acad. Sci. USA 117, 11459–11470 (2020).
Gonzalez-Sandoval, A. et al. Perinuclear anchoring of H3K9-methylated chromatin stabilizes induced cell fate in C. elegans embryos. Cell 163, 1333–1347 (2015).
Furukawa, K. LAP2 binding protein 1 (L2BP1/BAF) is a candidate mediator of LAP2-chromatin interaction. J. Cell Sci. 112, 2485–2492 (1999).
Mansharamani, M. & Wilson, K. L. Direct binding of nuclear membrane protein MAN1 to emerin in vitro and two modes of binding to barrier-to-autointegration factor. J. Biol. Chem. 280, 13863–13870 (2005).
Lee, K. K. et al. Distinct functional domains in emerin bind lamin A and DNA-bridging protein BAF. J. Cell Sci. 114, 4567–4573 (2001).
Samwer, M. et al. DNA cross-bridging shapes a single nucleus from a set of mitotic chromosomes. Cell 170, 956–972 e923 (2017).
Poleshko, A. et al. The human protein PRR14 tethers heterochromatin to the nuclear lamina during interphase and mitotic exit. Cell Rep. 5, 292–301 (2013).
Gorjanacz, M. et al. Caenorhabditis elegans BAF-1 and its kinase VRK-1 participate directly in post-mitotic nuclear envelope assembly. EMBO J. 26, 132–143 (2007).
Margalit, A. et al. Barrier to autointegration factor blocks premature cell fusion and maintains adult muscle integrity in C. elegans. J. Cell Biol. 178, 661–673 (2007).
Yang, M. & Yuan, Z. M. A novel role of PRR14 in the regulation of skeletal myogenesis. Cell Death Dis. 6, e1734 (2015).
Segura-Totten, M. & Wilson, K. L. BAF: roles in chromatin, nuclear structure and retrovirus integration. Trends Cell Biol. 14, 261–266 (2004).
Furukawa, K. et al. Barrier-to-autointegration factor plays crucial roles in cell cycle progression and nuclear organization in Drosophila. J. Cell Sci. 116, 3811–3823 (2003).
Homem, C. C., Reichardt, I., Berger, C., Lendl, T. & Knoblich, J. A. Long-term live cell imaging and automated 4D analysis of drosophila neuroblast lineages. PLoS ONE 8, e79588 (2013).
Ceron, J., Tejedor, F. J. & Moya, F. A primary cell culture of Drosophila postembryonic larval neuroblasts to study cell cycle and asymmetric division. Eur. J. Cell Biol. 85, 567–575 (2006).
Kwon, M. J. et al. Coiled-coil structure-dependent interactions between polyQ proteins and Foxo lead to dendrite pathology and behavioral defects. Proc. Natl Acad. Sci. USA 115, E10748–E10757 (2018).
Chung, C. G. et al. Golgi outpost synthesis impaired by toxic polyglutamine proteins contributes to dendritic pathology in neurons. Cell Rep. 20, 356–369 (2017).
Rosenberg, A. B. et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182 (2018).
Parekh, S., Ziegenhain, C., Vieth, B., Enard, W. & Hellmann, I. zUMIs—a fast and flexible pipeline to process RNA sequencing data with UMIs. Gigascience https://doi.org/10.1093/gigascience/giy059 (2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
McCarthy, D. J., Campbell, K. R., Lun, A. T. & Wills, Q. F. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186 (2017).
Lun, A. T., McCarthy, D. J. & Marioni, J. C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res 5, 2122 (2016).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Skinnider, M. A. et al. Cell type prioritization in single-cell data. Nat. Biotechnol. 39, 30–34 (2020).
Setty, M. et al. Characterization of cell fate probabilities in single-cell data with Palantir. Nat. Biotechnol. 37, 451–460 (2019).
Alpert, A., Moore, L. S., Dubovik, T. & Shen-Orr, S. S. Alignment of single-cell trajectories to compare cellular expression dynamics. Nat. Methods 15, 267–270 (2018).
Brunet Avalos, C., Maier, G. L., Bruggmann, R. & Sprecher, S. G. Single cell transcriptome atlas of the Drosophila larval brain. Elife https://doi.org/10.7554/eLife.50354 (2019).
Bier, E., Vaessin, H., Younger-Shepherd, S., Jan, L. Y. & Jan, Y. N. deadpan, an essential pan-neural gene in Drosophila, encodes a helix-loop-helix protein similar to the hairy gene product. Genes Dev. 6, 2137–2151 (1992).
Bowman, S. K. et al. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev. Cell 14, 535–546 (2008).
Zhu, S., Barshow, S., Wildonger, J., Jan, L. Y. & Jan, Y. N. Ets transcription factor Pointed promotes the generation of intermediate neural progenitors in Drosophila larval brains. Proc. Natl Acad. Sci. USA 108, 20615–20620 (2011).
Hirata, J., Nakagoshi, H., Nabeshima, Y. & Matsuzaki, F. Asymmetric segregation of the homeodomain protein Prospero during Drosophila development. Nature 377, 627–630 (1995).
Caussinus, E. & Hirth, F. Asymmetric stem cell division in development and cancer. Prog. Mol. Subcell. Biol. 45, 205–225 (2007).
Rives-Quinto, N. et al. Sequential activation of transcriptional repressors promotes progenitor commitment by silencing stem cell identity genes. Elife https://doi.org/10.7554/eLife.56187 (2020).
Corrales, M. et al. A single-cell transcriptomic atlas of complete insect nervous systems across multiple life stages. Neural Dev. 17, 8 (2022).
Dillon, N. et al. Single cell RNA-seq analysis reveals temporally-regulated and quiescence-regulated gene expression in Drosophila larval neuroblasts. Neural Dev. 17, 7 (2022).
Bannister, A. J. & Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 (2011).
Weng, R. & Cohen, S. M. Control of Drosophila Type I and Type II central brain neuroblast proliferation by bantam microRNA. Development 142, 3713–3720 (2015).
Torras-Llort, M. et al. A fraction of barrier-to-autointegration factor (BAF) associates with centromeres and controls mitosis progression. Commun. Biol. 3, 454 (2020).
Duan, T., Kitzman, S. C. & Geyer, P. K. Survival of Drosophila germline stem cells requires the chromatin-binding protein Barrier-to-autointegration factor. Development https://doi.org/10.1242/dev.186171 (2020).
Levine, M. S. & Holland, A. J. The impact of mitotic errors on cell proliferation and tumorigenesis. Genes Dev. 32, 620–638 (2018).
Hans, F. & Dimitrov, S. Histone H3 phosphorylation and cell division. Oncogene 20, 3021–3027 (2001).
Wolf, F. A. et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 20, 59 (2019).
Kohwi, M. & Doe, C. Q. Temporal fate specification and neural progenitor competence during development. Nat. Rev. Neurosci. 14, 823–838 (2013).
Skeath, J. B. & Thor, S. Genetic control of Drosophila nerve cord development. Curr. Opin. Neurobiol. 13, 8–15 (2003).
Bahrampour, S., Gunnar, E., Jonsson, C., Ekman, H. & Thor, S. Neural lineage progression controlled by a temporal proliferation program. Dev. Cell 43, 332–348 e334 (2017).
Jacobs, H. W., Keidel, E. & Lehner, C. F. A complex degradation signal in Cyclin A required for G1 arrest, and a C-terminal region for mitosis. EMBO J. 20, 2376–2386 (2001).
Royzman, I., Whittaker, A. J. & Orr-Weaver, T. L. Mutations in Drosophila DP and E2F distinguish G1-S progression from an associated transcriptional program. Genes Dev. 11, 1999–2011 (1997).
Ner, S. S. & Travers, A. A. HMG-D, the Drosophila melanogaster homologue of HMG 1 protein, is associated with early embryonic chromatin in the absence of histone H1. EMBO J. 13, 1817–1822 (1994).
Chalkley, G. E. et al. The transcriptional coactivator SAYP is a trithorax group signature subunit of the PBAP chromatin remodeling complex. Mol. Cell Biol. 28, 2920–2929 (2008).
Quinn, L. M. et al. Drosophila Hfp negatively regulates dmyc and stg to inhibit cell proliferation. Development 131, 1411–1423 (2004).
Homem, C. C. F. et al. Ecdysone and mediator change energy metabolism to terminate proliferation in Drosophila neural stem cells. Cell 158, 874–888 (2014).
Atwood, S. X. & Prehoda, K. E. aPKC phosphorylates Miranda to polarize fate determinants during neuroblast asymmetric cell division. Curr. Biol. 19, 723–729 (2009).
Prehoda, K. E. Polarization of Drosophila neuroblasts during asymmetric division. Cold Spring Harb. Perspect. Biol. 1, a001388 (2009).
Oon, C. H. & Prehoda, K. E. Asymmetric recruitment and actin-dependent cortical flows drive the neuroblast polarity cycle. Elife https://doi.org/10.7554/eLife.45815 (2019).
Rolls, M. M., Albertson, R., Shih, H. P., Lee, C. Y. & Doe, C. Q. Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J. Cell Biol. 163, 1089–1098 (2003).
Lee, C. Y., Robinson, K. J. & Doe, C. Q. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature 439, 594–598 (2006).
Shen, C. P., Jan, L. Y. & Jan, Y. N. Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell 90, 449–458 (1997).
Guo, L., Zhang, N. & Simpson, J. H. Descending neurons coordinate anterior grooming behavior in Drosophila. Curr. Biol. 32, 823–833 e824 (2022).
Hampel, S., Franconville, R., Simpson, J. H. & Seeds, A. M. A neural command circuit for grooming movement control. Elife 4, e08758 (2015).
McGuire, S. E., Mao, Z. & Davis, R. L. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci. STKE 2004, pl6 (2004).
Gallet, A. et al. The C-terminal domain of armadillo binds to hypophosphorylated teashirt to modulate wingless signalling in Drosophila. EMBO J. 18, 2208–2217 (1999).
Gallet, A., Erkner, A., Charroux, B., Fasano, L. & Kerridge, S. Trunk-specific modulation of wingless signalling in Drosophila by teashirt binding to armadillo. Curr. Biol. 8, 893–902 (1998).
Vishal, K. et al. Adult muscle formation requires drosophila moleskin for proliferation of wing disc-associated muscle precursors. Genetics 206, 199–213 (2017).
Rosenbloom, A. B. et al. beta-Catenin signaling dynamics regulate cell fate in differentiating neural stem cells. Proc. Natl Acad. Sci. USA 117, 28828–28837 (2020).
Tian, A., Benchabane, H., Wang, Z. & Ahmed, Y. Regulation of stem cell proliferation and cell fate specification by wingless/Wnt signaling gradients enriched at adult intestinal compartment boundaries. PLoS Genet 12, e1005822 (2016).
Pai, L. M., Orsulic, S., Bejsovec, A. & Peifer, M. Negative regulation of Armadillo, a Wingless effector in Drosophila. Development 124, 2255–2266 (1997).
Meister, P., Towbin, B. D., Pike, B. L., Ponti, A. & Gasser, S. M. The spatial dynamics of tissue-specific promoters during C. elegans development. Genes Dev. 24, 766–782 (2010).
Snyder, M. J. et al. Anchoring of heterochromatin to the nuclear lamina reinforces dosage compensation-mediated gene repression. PLoS Genet 12, e1006341 (2016).
Kafri, P. et al. Quantifying beta-catenin subcellular dynamics and cyclin D1 mRNA transcription during Wnt signaling in single living cells. Elife https://doi.org/10.7554/eLife.16748 (2016).
Fagotto, F. Looking beyond the Wnt pathway for the deep nature of beta-catenin. EMBO Rep. 14, 422–433 (2013).
Brand, A. H. & Livesey, F. J. Neural stem cell biology in vertebrates and invertebrates: more alike than different? Neuron 70, 719–729 (2011).
Homem, C. C., Repic, M. & Knoblich, J. A. Proliferation control in neural stem and progenitor cells. Nat. Rev. Neurosci. 16, 647–659 (2015).
Noctor, S. C., Flint, A. C., Weissman, T. A., Dammerman, R. S. & Kriegstein, A. R. Neurons derived from radial glial cells establish radial units in neocortex. Nature 409, 714–720 (2001).
Miyata, T., Kawaguchi, A., Okano, H. & Ogawa, M. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron 31, 727–741 (2001).
Franco, S. J. & Muller, U. Shaping our minds: stem and progenitor cell diversity in the mammalian neocortex. Neuron 77, 19–34 (2013).
Shen, Q. et al. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat. Neurosci. 9, 743–751 (2006).
Cox, J. L. et al. Banf1 is required to maintain the self-renewal of both mouse and human embryonic stem cells. J. Cell Sci. 124, 2654–2665 (2011).
Kuerbitz, J. et al. Loss of intercalated cells (ITCs) in the mouse amygdala of Tshz1 mutants correlates with fear, depression, and social interaction phenotypes. J. Neurosci. 38, 1160–1177 (2018).
Ragancokova, D. et al. TSHZ1-dependent gene regulation is essential for olfactory bulb development and olfaction. J. Clin. Invest. 124, 1214–1227 (2014).
Lorenzen, J. A. et al. Nuclear import of activated D-ERK by DIM-7, an importin family member encoded by the gene moleskin. Development 128, 1403–1414 (2001).
James, B. P., Bunch, T. A., Krishnamoorthy, S., Perkins, L. A. & Brower, D. L. Nuclear localization of the ERK MAP kinase mediated by Drosophila alphaPS2betaPS integrin and importin-7. Mol. Biol. Cell 18, 4190–4199 (2007).
Lanctot, A. A., Peng, C. Y., Pawlisz, A. S., Joksimovic, M. & Feng, Y. Spatially dependent dynamic MAPK modulation by the Nde1-Lis1-Brap complex patterns mammalian CNS. Dev. Cell 25, 241–255 (2013).
Wang, Y. et al. ERK inhibition rescues defects in fate specification of Nf1-deficient neural progenitors and brain abnormalities. Cell 150, 816–830 (2012).
Acknowledgements
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea, funded by the Ministry of Science and ICT, Republic of Korea (2022R1A4A2000703 and 2021R1A2C1003817 to S.B.L., 2021R1A6A1A10042944, 2021R1A6C101A390, and 2023R1A2C3004065 to J.K.K., 2016M3C7A1947307 and 2016M3C7A1904148 to H.L.) and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) and Korea Dementia Research Center (KDRC), funded by the Ministry of Health & Welfare and Ministry of Science and ICT, Republic of Korea (HU21C0027 to S.B.L.).
Author information
Authors and Affiliations
Contributions
B.S.K., M.H.H., M.J.K., D.G.C., J.K.K., and S.B.L. conceived the project. B.S.K., M.H.H., M.J.K., Y.J., and M.J.J. performed the Drosophila experiments and analyzed the data. D.G.C., E.S.P., S.K., Y.H.C., J.L., and J.K.K. performed the scRNA-seq and analyzed the data. B.S.K., M.H.H., M.J.K., D.G.C., K.L., M.T., K.J.Y., H.L., J.K.K., and S.B.L. interpreted the data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ko, B.S., Han, M.H., Kwon, M.J. et al. Baf-mediated transcriptional regulation of teashirt is essential for the development of neural progenitor cell lineages. Exp Mol Med 56, 422–440 (2024). https://doi.org/10.1038/s12276-024-01169-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s12276-024-01169-3