Abstract
Invariant natural killer T (iNKT) cells are thymus-generated innate-like αβ T cells that undergo terminal differentiation in the thymus. Such a developmental pathway differs from that of conventional αβ T cells, which are generated in the thymus but complete their functional maturation in peripheral tissues. Multiple subsets of iNKT cells have been described, among which IL-17-producing iNKT cells are commonly referred to as NKT17 cells. IL-17 is considered a proinflammatory cytokine that can play both protective and pathogenic roles and has been implicated as a key regulatory factor in many disease settings. Akin to other iNKT subsets, NKT17 cells acquire their effector function during thymic development. However, the cellular mechanisms that drive NKT17 subset specification, and how iNKT cells in general acquire their effector function prior to antigen encounter, remain largely unknown. Considering that all iNKT cells express the canonical Vα14-Jα18 TCRα chain and all iNKT subsets display the same ligand specificity, i.e., glycolipid antigens in the context of the nonclassical MHC-I molecule CD1d, the conundrum is explaining how thymic NKT17 cell specification is determined. Mapping of the molecular circuitry of NKT17 cell differentiation, combined with the discovery of markers that identify NKT17 cells, has provided new insights into the developmental pathway of NKT17 cells. The current review aims to highlight recent advances in our understanding of thymic NKT17 cell development and to place these findings in the larger context of iNKT subset specification and differentiation.
Similar content being viewed by others
Introduction
The antigen receptor of T cells, commonly referred to as the T cell receptor (TCR), is a defining feature of all T cells. The TCR is not only required for the generation of T cells in the thymus but also determines their antigen specificity, lineage choice, phenotype, and function, among other characteristics1. Consequently, the forced expression of a pre-rearranged TCR was found to determine the CD4 versus CD8 lineage fate as well as to control the choice of helper versus cytotoxic function and other characteristics of developing thymocytes2,3,4. Because the random, somatic recombination of TCR genes permits the generation of a vastly diverse TCR repertoire, conventional αβ T cells have access to a large pool of different TCR specificities to choose from for their TCR expression during thymic development5,6. In agreement, the TCRs of mature T cells comprise a highly diverse antigen repertoire7,8. Curiously, some T cells abstain from making use of the diverse TCR repertoire and instead employ a highly limited set of TCR chains for their generation. Because the cellular identity of T cells is imposed by the TCR, such oligoclonal T cells share common features in their antigen specificity and phenotype, and they can be pooled into specific T cell subsets with distinct features and functions.
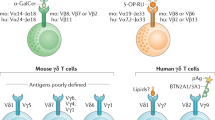
Invariant natural killer T (iNKT) cells are prime examples of such an oligoclonal T cell population, and they are distinct from conventional αβ T cells in several aspects related to their phenotype and effector functions9,10. Foremost, iNKT cells are heavily constrained in their TCR repertoire, as they all express the canonical Vα14-Jα18 TCRα chain in association with a TCRβ chain that is limited in its diversity, being Vβ2, Vβ7, or Vβ811,12. Importantly, the invariant Vα14-Jα18 TCRα chain restricts TCR binding to nonclassical MHC molecules, specifically to the nonclassical MHC-I-like molecule CD1d, that are bound to glycolipids13,14. The glycosphingolipid α-galactosylceramide (α-GalCer) is the prototypic antigen recognized by iNKT cells13,15, but other glycolipid analogs also have been identified to bind CD1d and be capable of activating iNKT cells14,16. Structural analyses revealed that the spatial interaction between glycolipid-loaded CD1d and the iNKT TCR is asymmetric and depends heavily on the invariant TCRα chain, with minimal contributions from the TCRβ chain, to form the binding interface17,18. Consequently, iNKT cells, which by definition all express the canonical Vα14-Jα18 TCRα chain, display the same MHC restriction and respond to the same antigens. As a corollary, all iNKT cells react to glycolipid-bound CD1d molecules and therefore can be identified by α-GalCer-loaded recombinant CD1d tetramers19,20,21. Thus, iNKT cells correspond to a subset of αβ T cells that are highly constrained in their TCR repertoire and thus share a limited antigen specificity.
Given that all iNKT cells express the same invariant TCRα chain and react to the same antigen, it could be assumed that the phenotypes and effector functions of individual iNKT cells would be uniform. Contrary to this expectation, the iNKT cell pool comprises many different subsets that are highly heterogeneous in their effector function, coreceptor expression, and tissue distribution22,23. Long-standing efforts to stratify these different iNKT cells have led to the proposal of different subsets of iNKT cells, either based on CD4 coreceptor expression24,25, developmental stage26, or effector function27,28. In the last case, iNKT cells could be classified into distinct subsets based on their cytokine production. Analogous to the Th1, Th2, and Th17 CD4 T helper subsets, iNKT cells that primarily express IFNγ, IL-4, or IL-17 have been referred to as NKT1, NKT2, and NKT17 cells, respectively27. These cytokine-based iNKT subsets can also be identified by their transcription factor expression profiles, whereby T-bet is exclusively expressed in NKT1 cells and RORγt is expressed in only NKT17 cells27,28. Similar to Th2 cells, NKT2 cells express high levels of GATA-3, but GATA-3 expression is not exclusive to NKT2 cells29. Thus, high-level expression of another transcription factor, the promyelocytic leukemia zinc-finger (PLZF) protein, is frequently used for more stringent identification of NKT2 cells27,30,31,32,33. Overall, iNKT cells comprise different functional subsets that display the same TCR specificity. As such, the holy grail for the understanding of iNKT subset specification is the elucidation of the mechanism through which the same TCR specificity can give rise to at least three distinct functional subsets; this is a highly active area of research10,23,34.
iNKT cells comprise only a small population of T cells, but they play disproportionally important roles in immune regulation and surveillance35,36,37. Specifically, IL-17-producing NKT17 cells have been implicated in both host defense against fungal infections and the pathogenesis of many autoimmune diseases, such as asthma and psoriasis, as well as graft-versus-host disease38,39,40. Nonetheless, how and when the developmental pathway of NKT17 cells diverts from those of other iNKT subsets remain mostly unknown, and why the frequency and number of NKT17 cells vary among different tissues and mouse strains is also unclear39,41,42,43. Here, we review and discuss recent progress in the field that has addressed these questions and provide a summary of different markers that identify NKT17 cells and how their expression is associated with NKT17 cell development and differentiation.
iNKT cell development in the thymus
The initial stages of iNKT cell generation parallel the development of conventional αβ T cells in the thymus. Both populations mostly arise from immature CD4+CD8+ double-positive (DP) thymocytes, at which stage they undergo positive selection44,45,46. Unlike that of conventional αβ T cells, however, the positive selection of iNKT cells is not mediated by thymic stromal cells in the cortex. Instead, iNKT cells are positively selected by glycolipid-loaded CD1d molecules that are expressed on thymocytes themselves and require homotypic costimulation by SLAM receptors (Fig. 1)46,47. Additionally, and unlike conventional αβ T cells that are generated by weak TCR engagement, iNKT cells are selected upon strong agonistic TCR signaling, which induces and requires the expression of the transcription factor early responsive gene 2 (Egr2)48,49,50. In agreement, the earliest iNKT-lineage cells in the thymus can be identified as Egr2+ cells that coexpress CD69 and bind to glycolipid-loaded CD1d tetramers48,49,50. These postselection immature iNKT cells are commonly referred to as stage 0 (ST0) cells or NKT0 cells26,51,52, and they express high levels of Nur77, indicative of the strong TCR signaling that mediates their generation (Fig. 1)53,54. Agonistic TCR signaling also highly upregulates the expression of PLZF, which is a critical nuclear factor required for the development and acquisition of effector function in iNKT cells30,31. ST0 iNKT cells express large amounts of CD24, a widely used marker for immature T cells55, but start downregulating CD24 expression upon further maturation that is associated with a c-Myc-dependent proliferative burst56,57. As a result of this expansion, three distinct subsets of iNKT cells, i.e., NKT1, NKT2, and NKT17, emerge; all of these subsets display a CD24lo phenotype, but they differ in their effector function and expression of PLZF (Fig. 1)27. A recent study identified a previously unappreciated precursor population among CD24lo iNKT cells, which is referred to as the NKT progenitor (NKTp) population and could give rise to all three iNKT subsets58. NKTp cells are marked by high-level expression of Egr2 and the chemokine receptors CCR7 and S1PR1 but lack effector function, even though they express large amounts of PLZF59. While the exact timing of iNKT subset specification remains unclear, the identification of the CD24lo NKTp population suggested that it could occur during or immediately after positive selection-induced proliferation.
In the thymic cortex, iNKT cells are positively selected by CD4+CD8+ double-positive (DP) thymocytes that present lipid antigens loaded on CD1d, a nonclassical MHC-I-like molecule. Selection into the iNKT cell lineage requires strong TCR signaling and homotypic interactions between signaling lymphocyte activation molecule (SLAM) family members. Post-selection iNKT cells are referred to as stage 0 or NKT0 cells, and they have high expression of Egr2, CD69, and CD24 but lack CCR7 expression. Immediately after positive selection, these progenitor cells undergo a c-Myc-dependent proliferative burst and proceed along a developmental pathway toward multipotent NKT cell precursors, designated as NKTp cells. These progenitor cells upregulate PLZF and CCR7 and downregulate CD24 expression before continuing terminal differentiation into three different iNKT subsets, namely, NKT1, NKT2, and NKT17 cells. NKT2 cells are GATA-3 positive and have the highest expression of PLZF, whereas NKT1 cells are T-bet positive and have the lowest PLZF expression. NKT17 cells are RORγt positive with intermediate PLZF expression.
Because PLZF is induced upon TCR signaling, the distinct amounts of PLZF among different iNKT subsets have led to the assumption that NKT2 cells receive the strongest TCR signal during development because they have the highest level of PLZF (PLZFhi), whereas NKT1 cells are selected by the weakest TCR signal and have the lowest level of PLZF (PLZFlo) expression31,32,60. NKT17 cells, on the other hand, contain an intermediate amount of PLZF (PLZFint), and they are proposed to be selected by intermediate-strength TCR signals (Fig. 1)31,32,60. However, how the difference in TCR signaling strength can be generated when all iNKT subsets express the same canonical TCR genes, even though they differ in their CDR3 sequences61, has not been fully explained. Specifically, what defines an intermediate-strength signal so that some iNKT precursors do not diverge into either the NKT1 or NKT2 lineage but become directed toward the NKT17 lineage is unclear. Nonetheless, it is evident that the different levels of PLZF are associated with the expression of iNKT subset-specific transcription factors, such that PLZFhi iNKT cells are GATA-3hi and PLZFint iNKT cells express RORγt, while PLZFlo iNKT cells exclusively express T-bet (Fig. 1)27,32,62. Consequently, it has been considered a key issue to understand how TCR signals are translated into the induction of different transcription factors and, specifically in the context of NKT17 cell differentiation, how this induction would lead to the NKT17-specific expression of RORγt.
NKT17 subset specification in the thymus
Among iNKT cells, RORγt is exclusively expressed in the NKT17 subset. Incidentally, RORγt is also highly expressed in DP thymocytes, which are the immediate precursors of immature iNKT cells45,46,63,64. In DP thymocytes, RORγt plays a critical role in cell survival by suppressing cellular metabolism65 and inducing antiapoptotic Bcl-xL expression66,67. Accordingly, RORγt-deficient mice fail to produce iNKT cells45 since their DP thymocytes cannot survive long enough to undergo successful rearrangement of the invariant Vα14-Jα18 chain that is encoded in the distal part of the TCRα gene45,68. In this context, the forced expression of transgenic Bcl-xL is sufficient to restore thymic iNKT cells, indicating that the primary role of RORγt in generating iNKT cells is to promote the survival of DP thymocytes45. Notably, such RORγt-deficient Bcl-xL-transgenic mice generate iNKT cells, but they still lack NKT17 cells. RORγt-deficient iNKT cells fail to become NKT17 cells because RORγt acts as the master regulator of NKT17 cell differentiation that equips these cells with a subset-specific phenotype and effector function45. Therefore, RORγt is expressed in both DP thymocytes and NKT17 cells but exerts dual functions in a cellular context-dependent manner.
Because RORγt is expressed in both DP thymocytes and NKT17 cells, it raises the question whether RORγt expression is ever turned off during the differentiation of DP thymocytes into NKT17 cells. Experimental data strongly support this hypothesis, and the current consensus is that positive selecting TCR signals terminate RORγt expression so that all iNKT subsets arise from a RORγt-negative precursor population. A recent study that employed a newly generated inducible Vα14-Jα18 TCR expression system reaffirmed this notion69. This genetically engineered mouse model permits monitoring of the development of a synchronized cohort of iNKT cells in vivo, and it revealed that positive selection of iNKT precursors terminates the expression of both the CD4 and CD8 coreceptors, rendering the cells CD4 and CD8 double-negative (CD4–CD8–), but also extinguishes their RORγt expression69. Thus, the immediate progeny of positively selected iNKT cells appear to be RORγt–CD4–CD8– CD1d-tetramer-positive cells. These postselection immature iNKT cells then undergo induction of the transcription factor ThPOK and pass through a CD4+ stage that is common to all iNKT cells, after which they undergo lineage specification into different iNKT subsets51,59,69. Collectively, these results argue for dynamic regulation of RORγt expression during NKT17 cell differentiation, in which RORγt expression is transiently downregulated upon positive selection but then reinduced upon NKT17 lineage commitment.
Such a developmental trajectory of NKT17 cells necessitates a molecular explanation of how RORγt is re-expressed in NKT17 cells. Because of their IL-17-producing characteristics, NKT17 cells have frequently been compared to IL-17-producing CD4+ Th17 cells39,70. Th17 cells can be generated from naive CD4 T cells by TCR activation in the presence of the cytokines TGF-β and IL-671. Thus, it seems feasible that the same cytokine combination would also drive RORγt induction and NKT17 cell differentiation. This presumption, however, is not quite correct. While NKT17 cells express high levels of TGF-β receptors and show markedly elevated phosphorylation of SMAD2/3 when freshly extracted from the thymus69,72, IL-6 is not required for the generation of NKT17 cells72,73,74 (Fig. 2). Moreover, in humans, in which NKT17 cells were first described as CD161+ iNKT cells, the differentiation and acquisition of effector function of NKT17 cells depend not only on TGF-β but also on IL-1β and IL-2375. Importantly, the IL-23 receptor is highly and selectively expressed on NKT17 cells, so it has been considered a prominent marker for the NKT17 subset of iNKT cells73,76, whereby IL-23 receptor signaling possibly plays a critical role in NKT17 lineage commitment rather than maintenance77. Overall, TGF-β is a common denominator for both mouse and human NKT17 cells as well as a common denominator between Th17 and NKT17 cells, but there are variations in the requirements for costimulatory cytokines between these cells. In this regard, it remains unclear how the coordinated effects of TGF-β and other cytokines contribute to the induction of RORγt and the specification of the NKT17 subset. In TGF-β signaling, for example, conditional deletion of SMAD4 but not TRIM33 was shown to impair NKT17 cell generation, although the downstream target genes of SMAD activation have yet to be identified72. The same study showed that constitutive expression of TGF-β promoted the survival and accumulation of NKT17 cells in peripheral lymph nodes but did not affect NKT17 cell generation in the thymus72. Thus, the cytokine requirements for NKT17 cells can differ depending on the tissue environment, and the downstream targets of NKT17-specifying cytokines remain to be determined.
NKT17 cells can be identified by several cell-surface markers. CD138 (syndecan-1) is a heparan sulfate proteoglycan that is specifically expressed on NKT17 cells. CD138 interacts with extracellular matrix proteins, chemokines, cytokines, growth factors and integrins, among others, but its specific role, if any, in NKT17 cell biology remains unknown. CCR6 is a chemokine receptor associated with NKT17 cells that promotes their migration into tissues rich in its ligand CCL20, such as the skin, lungs, and lymph nodes. DR3 is a TNF receptor superfamily member that is expressed mostly by the NKT17 subset among thymic iNKT cells. Upon interaction with its endogenous ligand, TL1A, DR3 acts as a costimulatory molecule in context of the α-galactosylceramide-mediated activation of NKT17 cells. IL-23 receptor (IL-23R) is exclusively expressed on NKT17 cells, and its activation triggers IL-17A secretion. TGF-βR is a cytokine receptor for the multifunctional cytokine TGF-β. Analogous to its role in the Th17 cell polarization of CD4+ T cells, TGF-βR ligation augments NKT17 cell specification in iNKT cells. mTEC, medullary thymic epithelial cells.
Nuclear factors controlling NKT17 subset specification
The distinct TGF-β signature in NKT17 cells suggests that cytokine signaling contributes to iNKT subset specification, either in addition to or in concert with TCR signaling69,72. In fact, it is likely that positive selection and lineage specification of iNKT cells are disparate events, as is the case for the thymic generation of conventional CD4 and CD8αβ T cells78. In this setting, TCR signals would be mostly required for positive selection, and cytokine signals would determine the subset identity, as recently proposed69. However, it is unclear how only some iNKT cells would respond to a specific cytokine, such as TGF-β, to commit to a particular subset, while other iNKT cells are nonresponsive to the same cytokine. Whether the strength and/or kinetics of the preceding or concomitant TCR signal play a role in this process is a possibility that can be tested. However, a clear distinction between or overlap of the cytokine versus TCR contributions in iNKT subset differentiation has yet to be established. As such, assessing how the downstream signaling pathways of TCR and cytokine signaling intersect to establish iNKT subset identity remains an area of active research.
NKT17 cells express intermediate levels of the transcription factors PLZF and Egr2, whose abundances correlate with the strength of TCR signaling32,50,60. Hypothetically, intermediate-strength TCR signals could be necessary to transition immature iNKT cells into a TGF-β signaling-permissive state so that they can induce RORγt in response to TGF-β. Weaker or stronger TCR signals, on the other hand, would keep immature iNKT cells refractory to TGF-β and thus prevent the induction of RORγt. Such a scenario implies that RORγt expression would be actively suppressed in non-NKT17 lineage-committed iNKT cells and that either PLZF or Egr2 could be involved in this process. However, both PLZF- and Egr2-deficient mice fail to generate mature iNKT cells30,31,49,50, making it difficult to test the roles of these transcription factors in iNKT subset specification.
Another prominent transcription factor whose expression is associated with TCR signal strength is the zinc finger transcription factor ThPOK79. ThPOK is absent in immature thymocytes but highly upregulated upon strong and persistent TCR signaling80,81. In agreement, CD4-lineage T cells, which require strong/persistent TCR signaling for their generation, express high levels of ThPOK, and their differentiation depends on ThPOK80,82. Notably, ThPOK is also highly expressed in iNKT cells, which is consistent with their requirement for strong agonistic TCR signaling53,83,84. However, unlike conventional CD4 T cells, which are virtually absent in ThPOK-deficient mice80,82,85, iNKT cells still develop in the absence of ThPOK, indicating that ThPOK presumably plays distinct roles in iNKT cells versus conventional CD4 T cells83,86. In this regard, ThPOK suppresses cytokine receptor signaling by controlling the expression of SOCS family molecules87, so ThPOK can bridge TCR signaling with cytokine signaling in iNKT cell differentiation. The iNKT subset analysis of ThPOK-deficient iNKT cells bolstered this possibility, noting that mice with a missense mutation in ThPOK or germline deficiency in ThPOK displayed dramatic increases in the frequency and number of thymic NKT17 cells83,88,89. ThPOK deficiency was also shown to promote NKT17 cell differentiation in the spleen and liver compared to control expression88,89. Conversely, the forced expression of ThPOK was found to potently suppress RORγt expression and the differentiation of IL-17-producing iNKT cells89. Thus, ThPOK acts as a suppressor of RORγt expression during iNKT subset specification, revealing a new layer of control in NKT17 cell generation. Whether ThPOK suppresses RORγt expression directly by interfering with its transcription or indirectly by modulating cytokine responsiveness or the expression of other factors is an important question that needs to be addressed.
The runt-family transcription factor Runx3 is a major target of ThPOK90. Because ThPOK suppresses Runx3 expression87,90,91, under normal circumstances, the expression of Runx3 and ThPOK is mutually exclusive92. As a corollary, Runx3 is mostly absent when ThPOK is expressed, as is the case in both CD4 cells and iNKT cells85,93. Interestingly, ThPOK-deficient iNKT cells show marked induction of Runx3 that is associated with ectopic CD8 coreceptor expression and altered effector function93. In this regard, it could be either the absence of ThPOK or the increased expression of Runx3 that promotes RORγt expression and NKT17 differentiation in ThPOK-deficient mice. Further experimental data and new mouse models, such as iNKT-specific overexpression of Runx3, are necessary to discriminate these possibilities. Nonetheless, a requirement for Runx3 in iNKT cell generation could be excluded based on the observation that Runx3-conditional knockout mice that lack Runx3 are mostly unaffected in their thymic iNKT cell differentiation93. Thus, unlike ThPOK, Runx3 is not a major contributor to the differentiation of iNKT cells.
In addition to Runx3, the Runx family members include Runx1 and Runx294. However, only Runx1 and Runx3 are expressed in lymphocytes85,93,95. In contrast to Runx3, which is primarily expressed in CD8 T cells and NK cells, Runx1 is highly expressed in CD4 T cells and iNKT cells93,96, suggesting that Runx1 rather than Runx3 could play a role in iNKT cells. Indeed, Runx1 deficiency in preselection thymocytes results in a complete block of iNKT cell development at the immature DP stage45, while conditional deletion of Runx1 with PLZF-Cre severely impairs the functional maturation of positively selected iNKT cells97. These findings support Runx1 as a nonredundant requirement in iNKT cell generation and differentiation. Since RORγt is suppressed by ThPOK, which in turn is antagonized by Runx3, Runx1 could potentially interfere with RORγt expression. In fact, Runx1-mediated activation of RORγt was previously demonstrated in IL-17-producing Th17 CD4 T cells98, and it is reasonable to assume that a similar pathway also operates in iNKT cells. In agreement, analyses of Runx1-deficient mice have shown that their iNKT cells exhibited significant reductions in the frequency and number of NKT17 cells and that cytokine production was selectively impaired in the NKT17 subset but not in NKT1 or NKT2 cells. Collectively, these results strongly suggest that Runx1 is a positive regulator of the transcriptional program that governs NKT17 cells.
Mechanistically, however, Runx1 could be controlling not only the expression of RORγt but also that of other genes associated with NKT17 cell differentiation. In this regard, it was interesting to find that the expression of c-Maf, a transcription factor that is highly enriched in NKT17 cells99, was markedly reduced in Runx1-deficient iNKT cells. Runx1 deficiency also induced a significant decrease in the expression of BATF, a transcription factor that promotes the in vitro differentiation of CD4+ Th17 cells100, resulting in a dramatic loss of IL-17 production in NKT17 cells. Reciprocally, BATF overexpression has been shown to skew the iNKT subset composition toward NKT17 cells, further unraveling the complex regulatory pathway of NKT17 cell differentiation that is controlled by Runx1 and associated transcription factors. Evidently, there is an ever-growing body of regulatory factors identified to be involved in the subset differentiation of NKT17 cells, and they include the transcriptional repressor NKAP101, which is specifically required for the differentiation of NKT17 but not NKT1 or NKT2 cells, as illustrated by the marked paucity of NKT17 cells in NKAP-deficient mice101. The mRNA-binding protein Roquin102 and the transcription factor Bcl11b103, on the other hand, are negative regulators of NKT17 cell generation, that could play yet to be assessed roles in RORγt expression at the posttranscriptional or transcriptional level. Connecting these factors into a comprehensive network is a daunting task, which will require comprehensive pathway analyses together with in-depth bioinformatic approaches.
Surface markers that identify NKT17 cells
The term NKT17 cells was coined in the seminal study by Michel and colleagues104, in which IL-17-producing iNKT cells were identified in the spleen, liver, and lungs as NK1.1-negative iNKT cells. Thus, starting early on, the phenotype of iNKT cells, such as expressing the surface marker NK1.1 expression, has been closely associated with effector function. However, not all NK1.1-negative iNKT cells are NKT17 cells. In fact, the NK1.1-negative iNKT population contains two distinct subsets that can be distinguished based on CD44 expression26,70. Because CD44 expression is thought to be an acquired trait upon thymic maturation, CD44–NK1.1– thymic iNKT cells are commonly referred to as stage 1 (ST1), while CD44+NK1.1– thymic iNKT cells correspond to stage 2 (ST2). iNKT cells that express both CD44 and NK1.1 (CD44+NK1.1+) are considered to have undergone terminal differentiation, and they are known as stage 3 (ST3) iNKT cells9,26,70. Detailed analyses of their functional characteristics place IFNγ-producing NKT1 cells into the ST3 compartment. In contrast, NKT17 cells are excluded from the ST3 subset, and they correspond mostly to ST2 cells. However, not all ST2 cells are NKT17 cells, as this population is heavily contaminated with NKT2 cells105. Therefore, alternative or additional markers are required to identify NKT17 cells.
Based on CD4 coreceptor expression, iNKT cells can be either CD4-positive or CD4-negative106. IL-17-producing iNKT cells are mostly found in the CD4-negative compartment, so NKT17 cells have been proposed to be phenotypically CD4–NK1.1– cells76. In fact, the lack of both CD4 and NK1.1 expression has been employed for a long time as a surrogate marker for NKT17 cells48,76,107. Alternatively, the IL-2/IL-15 receptor β-chain, i.e., CD122, is exclusively expressed on NKT1 cells, and combined with the observation that all NKT2 cells express CD4, the use of CD122 and CD4 can discriminate the three major subsets of iNKT cells. Accordingly, NKT1 cells are CD122+ and NKT2 cells are CD4+CD122–, while NKT17 cells are identified as CD122–CD4– double-negative (DN) cells27,48. Accordingly, the CD122–CD4– DN compartment corresponds mostly to RORγt+ iNKT cells108.
However, further in-depth studies of IL-17 production have revealed that a significant fraction of NKT17 cells are also found among CD4+ iNKT cells. Such CD4-expressing NKT17 cells have been reported in the thymus and lymph nodes of both BALB/c mice and C57BL/6 mice29, and ~10% of NKT17 cells residing in the mesenteric lymph nodes are CD4-positive72. The relative frequency of CD4-positive NKT17 cells among total NKT17 cells in different tissues is still debated, but a lack of CD4 expression alone cannot be employed to identify all NKT17 cells29. Consequently, alternative approaches making use of markers other than CD4 to identify NKT17 cells have been reported.
A powerful tool for NKT17 cell identification was devised using the differential expression of ICOS (CD278) and the activation-associated glycoform of CD43 (CD43HG) in iNKT cells29. Accordingly, NKT1 cells are identified as CD43– ICOS-low cells, and NKT2 cells are identified as CD43-intermediate ICOS+ cells, whereas NKT17 cells are CD43+ ICOS-high cells29. To confirm correct identification, iNKT subsets stratified by ICOS versus CD43HG staining were assessed for intracellular T-bet and RORγt staining and found to match their expected subset characteristics29. Along these lines, NKT2 cells that were purified based on CD43 and ICOS expression produced negligible levels of IL-17A, affirming that these markers accurately identified and excluded specific subsets29. Recently, a different set of surface markers was employed to identify NKT17 cells in FVB/N mice; NKT1 cells were first excluded from total iNKT cells based on their lack of PD-1 and ICOS expression and then gated on CD4-negative but CD27+ cells among the remaining PD-1+ ICOS+ iNKT cells38. Thus, using a combination of markers and gating strategies permits correct discrimination of individual iNKT subsets. Such an approach has turned out to be highly effective, but it is also cumbersome and complicated because multiple markers are required to identify the desired iNKT subset.
Accordingly, it would be more effective if the target iNKT subset could be identified by a single marker that is exclusively expressed by that particular iNKT cell population. CD122 corresponds to such a marker for NKT1 cells because it is expressed by only this specific iNKT subset and is incidentally also required for IL-15 signaling, which induces NKT1-specific T-bet expression109,110. In this regard, surface markers that are related to the function or a developmental requirement of a given iNKT subset could serve as faithful markers to identify that specific subset among other iNKT cells.
iNKT subsets are distinct in their tissue tropism and residency, given their differential chemokine receptor and cell adhesion molecule expression10,41,43. As such, NKT17 cells are especially enriched in the skin and lungs27,43,104. The chemokine receptor CCR6 is a major chemokine receptor in Th17 cells111 and NKT17 cells107, which could explain the enrichment of these subsets in the skin epithelium and mucosal tissues in which CCL20, the ligand for CCR6, is highly expressed (Fig. 2)112. In agreement, CCR6 mRNA transcripts were found exclusively in NKT17 cells among different iNKT subsets107. However, surface CCR6 expression is heterogeneous among NKT17 cells72, indicating that CCR6 can mark NKT17 cells but that not all NKT17 cells are necessarily CCR6 positive72. Such heterogeneity in CCR6 expression renders this chemokine receptor an unpredictable marker for NKT17 cells. Likewise, neuropilin-1, CD103, and CD121a have been proposed as promising candidates to identify NKT17 cells72. However, all of them were later found to be either heterogeneously expressed among NKT17 cells or not entirely specific to the NKT17 subset. Thus, a marker that could identify NKT17 cells has not been successfully identified among cytokine and chemokine receptors or cell adhesion molecules.
Serendipitously, CD138 (Syndecan-1) was recently discovered to be an NKT17-specific surface marker (Fig. 2)113. In support of this notion, only CD138+ iNKT cells produce IL-17 among all iNKT cells113. CD138 is a transmembrane heparan sulfate proteoglycan that is highly expressed on epithelial cells and plasma cells114. In T cells and thymocytes, CD138 is expressed on only a subset of mature CD4, CD8 DN thymocytes, among which the majority of CD138+ cells were found to be NKT17 cells, with a minor population of γδ T cells108. Currently, the role of CD138 in NKT17 cells is not fully understood. CD138 usually interacts with cell matrix proteins, cytokines, and growth factors114, so it could play regulatory roles in the tissue distribution, proliferation, or activation of NKT17 cells. However, CD138-deficient Sdc1–/– mice do not show any defects in the generation of NKT17 cells, and IL-17 production by Sdc1–/– NKT17 cells is also unimpaired108,113. Rather, both the frequency and number of thymic NKT17 cells are modestly increased, suggesting that CD138 does not play a major role in NKT17 cells and that even if it did, it would play a negative regulatory role108. To this extent, CD138 expression is mostly considered as a surface marker for NKT17 cells, with no clear biological function in NKT17 cells yet elucidated.
The ongoing search for a functional marker recently yielded the surprising discovery of Death Receptor-3 (DR3, also APO-3) as an NKT17 subset-specific molecule115 (Fig. 2). DR3 is a member of the TNF receptor superfamily that is activated by its only known ligand, TL1A, to trigger proinflammatory and apoptotic downstream signaling116. TL1A is primarily produced by antigen-presenting cells, such as dendritic cells and macrophages, but also by thymic medullary epithelial cells116, suggesting a potential role for the DR3-TL1A signaling pathway in T cell development (Fig. 2). Along these lines, DR3 was previously reported to be highly expressed on Foxp3+ CD4+ T regulatory cells (Tregs) to promote their expansion and partly contribute to their effector function117. As such, DR3 ligation with agonistic antibodies was shown to result in a dramatic expansion of Foxp3+ Treg cells in vivo117 and to ameliorate disease severity in acute GVHD settings117. A mouse model of constitutive TL1A expression also exhibited expansion of the pool of DR3-expressing cells, identifying group 2 innate lymphoid cells (ILC2s) as a target of TL1A118. Here, the excessive production of TL1A resulted in a significant increase in the ILC2 population, concomitant with an IL-13-mediated allergic immune response118. The expression of DR3 on thymic iNKT cells, specifically on NKT17 cells, however, had not been documented until recently115. Notably, NKT17-specific DR3 expression was mostly limited to NKT17 cells in the thymus, as DR3 expression is rather promiscuous on iNKT cells in peripheral tissues. While DR3 remains highly expressed on NKT17 cells, there is a substantial amount of DR3 expression on iNKT subsets in the lymph nodes and lungs, among other tissues115. However, what drives DR3 expression on peripheral iNKT cells that lack RORγt remains to be determined.
Because of the subset-specific expression of some cytokine receptors, such as CD122 for NKT1 cells and IL-17RB for NKT2 and NKT17 cells119,120, it is not surprising that there would be a proprietary cytokine receptor for NKT17 cells, which turned out to be DR3115. The molecular basis of NKT17-specific DR3 expression could be traced back to RORγt, which was sufficient to induce DR3 expression on conventional αβ T cells and non-NKT17 cells in thymocytes of RORγt-transgenic mice115. These results suggest that DR3 expression is a direct target of RORγt. In agreement, it was previously reported that DR3 is specifically expressed on Th17 cells, the only T helper cell subset that expresses RORγt121. Regarding the requirement for and effect of DR3 in NKT17 cells, however, we are only beginning to understand the role of DR3. While in vitro stimulation with agonistic anti-DR3 antibodies is sufficient to induce the early activation marker CD69, DR3 ligation alone fails to induce a sufficiently strong response to boost IL-17 production. Instead, DR3 acts more like a costimulatory molecule, as it was found to dramatically amplify the effect of α-GalCer stimulation and to increase IL-17 production and cell proliferation of thymic NKT17 cells115. Therefore, DR3 represents a new class of costimulatory molecules on thymic NKT17 cells that can serve as both a marker and a trigger of a specific iNKT subset.
Perspectives
iNKT cells undergo terminal differentiation in the thymus, upon which they egress into peripheral organs to establish tissue residency9,10,43,59. Generally, the subset identity of iNKT cells is considered developmentally fixed, so that NKT17 cells do not further differentiate into NKT1 cells and NKT2 cells or vice versa in the periphery27. However, the iNKT subset composition in the thymus is strikingly different from that in peripheral organs, and it also varies among different organs10. In this regard, NKT1 cells are highly enriched in the liver, while NKT17 cells accumulate in the lymph nodes and lungs43. Several distinct but not mutually exclusive models have been proposed to explain the tissue-specific distributions of individual iNKT subsets. A straightforward explanation would be that the thymic export and tissue tropism of iNKT cells differ among subsets. For example, some iNKT cells could efficiently exit the thymus and migrate to their target tissues, while others would be impaired in thymic egress and become thymus resident. In this regard, NKT2 cells express large amounts of CD69, which retains them in the thymus33, whereas NKT1 and NKT17 cells express the chemokine receptors CXCR3 and CCR6, respectively, which could attract them into peripheral tissues where their ligands are highly expressed48. CXCR6 is a chemokine receptor that is important for the survival and maintenance of iNKT cells in the liver, whereas the factors required for iNKT cell homing to the lungs have yet to be determined10,41. Thus, differences in thymic egress and tissue tropism could cause the differences in the iNKT subset composition between the thymus and peripheral organs.
Another attractive hypothesis is that each tissue environment provides unique survival signals that are tailored to each iNKT subset, resulting in the preferential survival and accumulation of a particular subset. In this regard, NKT17 cells reportedly prefer IL-7 over IL-15 signaling for their survival42,110,122, and this IL-7 dependence would cause enrichment in the lymph nodes, where IL-7 is abundantly expressed123. Why NKT17 cells would be more responsive to and dependent on IL-7 than the other iNKT subsets is not fully understood. A recent study demonstrated that forkhead box protein O1 (FoxO1) plays a critical role in NKT1 and NKT2 cells but not in NKT17 cells124. Notably, expression of the IL-7 receptor, which is highly abundant on all iNKT cells, is driven by FoxO1 in NKT1 and NKT2 cells but not in NKT17 cells. Instead, it turned out that it is RORγt which promotes IL-7 receptor expression on NKT17 cells124.
Overall, RORγt remains a critical master regulator of NKT17 cells that guides not only the generation and differentiation but also the survival and homeostasis of this iNKT subset. The observation that RORγt also controls the expression of surface molecules that mark NKT17 cells provides further evidence that the phenotype and function of NKT17 cells are closely associated with each other. Considering that the role of NKT17 cells is still being unraveled, it is important to decipher which NKT17 markers are functional and contribute to NKT17 biology, and which molecules are passenger markers with no apparent function. Further synthesis of this information will provide us with a better understanding of NKT17 cells in immunity.
References
Starr, T. K., Jameson, S. C. & Hogquist, K. A. Positive and negative selection of T cells. Annu. Rev. Immunol. 21, 139–176 (2003).
Teh, H. S. et al. Thymic major histocompatibility complex antigens and the αβ T-cell receptor determine the CD4/CD8 phenotype of T cells. Nature 335, 229–233 (1988).
Singer, A., Adoro, S. & Park, J. H. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat. Rev. Immunol. 8, 788–801 (2008).
Kaye, J. et al. Selective development of CD4+ T cells in transgenic mice expressing a class II MHC-restricted antigen receptor. Nature 341, 746–749 (1989).
Davis, M. M. & Bjorkman, P. J. T-cell antigen receptor genes and T-cell recognition. Nature 334, 395–402 (1988).
Goncalves, P. et al. A new mechanism shapes the naive CD8(+) T cell repertoire: the selection for full diversity. Mol. Immunol. 85, 66–80 (2017).
Vrisekoop, N., Monteiro, J. P., Mandl, J. N. & Germain, R. N. Revisiting thymic positive selection and the mature T cell repertoire for antigen. Immunity 41, 181–190 (2014).
de Greef, P. C. et al. The naive T-cell receptor repertoire has an extremely broad distribution of clone sizes. eLife 9, e49900 (2020).
Bendelac, A., Savage, P. B. & Teyton, L. The biology of NKT cells. Annu. Rev. Immunol. 25, 297–336 (2007).
Crosby, C. M. & Kronenberg, M. Tissue-specific functions of invariant natural killer T cells. Nat. Rev. Immunol. 18, 559–574 (2018).
Dellabona, P., Padovan, E., Casorati, G., Brockhaus, M. & Lanzavecchia, A. An invariant Vα24-JαQ/Vβ11 T cell receptor is expressed in all individuals by clonally expanded CD4-8- T cells. J. Exp. Med. 180, 1171–1176 (1994).
Lantz, O. & Bendelac, A. An invariant T cell receptor α chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4-8- T cells in mice and humans. J. Exp. Med. 180, 1097–1106 (1994).
Kawano, T. et al. CD1d-restricted and TCR-mediated activation of Vα14 NKT cells by glycosylceramides. Science 278, 1626–1629 (1997).
Rossjohn, J., Pellicci, D. G., Patel, O., Gapin, L. & Godfrey, D. I. Recognition of CD1d-restricted antigens by natural killer T cells. Nat Rev. Immunol. 12, 845–857 (2012).
Burdin, N. et al. Selective ability of mouse CD1 to present glycolipids: α-galactosylceramide specifically stimulates Vα14+ NK T lymphocytes. J. Immunol. 161, 3271–3281 (1998).
Zajonc, D. M. & Girardi, E. Recognition of microbial glycolipids by natural killer T cells. Front. Immunol. 6, 400 (2015).
Mallevaey, T. et al. A molecular basis for NKT cell recognition of CD1d-self-antigen. Immunity 34, 315–326 (2011).
Kjer-Nielsen, L. et al. A structural basis for selection and cross-species reactivity of the semi-invariant NKT cell receptor in CD1d/glycolipid recognition. J. Exp. Med. 203, 661–673 (2006).
Benlagha, K., Weiss, A., Beavis, A., Teyton, L. & Bendelac, A. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J. Exp. Med. 191, 1895–1903 (2000).
Matsuda, J. L. et al. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J. Exp. Med. 192, 741–754 (2000).
Karadimitris, A. et al. Human CD1d-glycolipid tetramers generated by in vitro oxidative refolding chromatography. Proc. Natl Acad. Sci. USA 98, 3294–3298 (2001).
Krovi, S. H. & Gapin, L. Invariant natural killer T cell subsets-more than just developmental intermediates. Front. Immunol. 9, 1393 (2018).
Wang, H. & Hogquist, K. A. How lipid-specific T cells become effectors: the differentiation of iNKT subsets. Front. Immunol. 9, 1450 (2018).
Bendelac, A., Rivera, M. N., Park, S. H. & Roark, J. H. Mouse CD1-specific NK1 T cells: development, specificity, and function. Annu. Rev. Immunol. 15, 535–562 (1997).
Crowe, N. Y. et al. Differential antitumor immunity mediated by NKT cell subsets in vivo. J. Exp. Med. 202, 1279–1288 (2005).
Benlagha, K., Kyin, T., Beavis, A., Teyton, L. & Bendelac, A. A thymic precursor to the NK T cell lineage. Science 296, 553–555 (2002).
Lee, Y. J., Holzapfel, K. L., Zhu, J., Jameson, S. C. & Hogquist, K. A. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat. Immunol. 14, 1146–1154 (2013).
Kwon, D. I. & Lee, Y. J. Lineage differentiation program of invariant natural killer T cells. Immune Netw. 17, 365–377 (2017).
Cameron, G. & Godfrey, D. I. Differential surface phenotype and context-dependent reactivity of functionally diverse NKT cells. Immunol. Cell Biol. 96, 759–771 (2018).
Kovalovsky, D. et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat. Immunol. 9, 1055–1064 (2008).
Savage, A. K. et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29, 391–403 (2008).
Park, J. Y., DiPalma, D. T., Kwon, J., Fink, J. & Park, J. H. Quantitative difference in PLZF protein expression determines iNKT lineage fate and controls innate CD8 T cell generation. Cell Rep. 27, 2548–2557 e2544 (2019).
Kimura, M. Y. et al. CD69 prevents PLZF(hi) innate precursors from prematurely exiting the thymus and aborting NKT2 cell differentiation. Nat. Commun. 9, 3749 (2018).
Bharadwaj, N. S. & Gumperz, J. E. Harnessing invariant natural killer T cells to control pathological inflammation. Front. Immunol. 13, 998378 (2022).
Van Kaer, L. & Wu, L. Therapeutic potential of invariant natural killer T cells in autoimmunity. Front. Immunol. 9, 519 (2018).
Godfrey, D. I., Le Nours, J., Andrews, D. M., Uldrich, A. P. & Rossjohn, J. Unconventional T cell targets for cancer immunotherapy. Immunity 48, 453–473 (2018).
McEwen-Smith, R. M., Salio, M. & Cerundolo, V. The regulatory role of invariant NKT cells in tumor immunity. Cancer Immunol. Res. 3, 425–435 (2015).
Maas-Bauer, K. et al. Invariant natural killer T-cell subsets have diverse graft-versus-host-disease-preventing and antitumor effects. Blood 138, 858–870 (2021).
Tsagaratou, A. Unveiling the regulation of NKT17 cell differentiation and function. Mol. Immunol. 105, 55–61 (2019).
Hu, Y. et al. Dysregulated peripheral invariant natural killer T cells in plaque psoriasis patients. Front. Cell. Dev. Biol. 9, 799560 (2021).
Slauenwhite, D. & Johnston, B. Regulation of NKT cell localization in homeostasis and infection. Front. Immunol. 6, 255 (2015).
Webster, K. E. et al. IL-17-producing NKT cells depend exclusively on IL-7 for homeostasis and survival. Mucosal Immunol. 7, 1058–1067 (2014).
Lee, Y. J. et al. Tissue-specific distribution of iNKT cells impacts their cytokine response. Immunity 43, 566–578 (2015).
Bendelac, A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J. Exp. Med. 182, 2091–2096 (1995).
Egawa, T. et al. Genetic evidence supporting selection of the Vα14i NKT cell lineage from double-positive thymocyte precursors. Immunity 22, 705–716 (2005).
Gapin, L., Matsuda, J. L., Surh, C. D. & Kronenberg, M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat. Immunol. 2, 971–978 (2001).
Griewank, K. et al. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity 27, 751–762 (2007).
Georgiev, H., Ravens, I., Benarafa, C., Forster, R. & Bernhardt, G. Distinct gene expression patterns correlate with developmental and functional traits of iNKT subsets. Nat. Commun. 7, 13116 (2016).
Lazarevic, V. et al. The gene encoding early growth response 2, a target of the transcription factor NFAT, is required for the development and maturation of natural killer T cells. Nat. Immunol. 10, 306–313 (2009).
Seiler, M. P. et al. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat. Immunol. 13, 264–271 (2012).
Benlagha, K., Wei, D. G., Veiga, J., Teyton, L. & Bendelac, A. Characterization of the early stages of thymic NKT cell development. J. Exp. Med. 202, 485–492 (2005).
Pellicci, D. G. et al. A natural killer T (NKT) cell developmental pathway iInvolving a thymus-dependent NK1.1(-)CD4(+) CD1d-dependent precursor stage. J. Exp. Med. 195, 835–844 (2002).
Moran, A. E. et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 208, 1279–1289 (2011).
Kumar, A. et al. Nur77 controls tolerance induction, terminal differentiation, and effector functions in semi-invariant natural killer T cells. Proc. Natl Acad. Sci. USA 117, 17156–17165 (2020).
Hough, M. R., Takei, F., Humphries, R. K. & Kay, R. Defective development of thymocytes overexpressing the costimulatory molecule, heat-stable antigen. J. Exp. Med. 179, 177–184 (1994).
Dose, M. et al. Intrathymic proliferation wave essential for Vα14+ natural killer T cell development depends on c-Myc. Proc. Natl Acad. Sci. USA 106, 8641–8646 (2009).
Mycko, M. P. et al. Selective requirement for c-Myc at an early stage of V(alpha)14i NKT cell development. J. Immunol. 182, 4641–4648 (2009).
Lee, Y. J. et al. Lineage-specific effector signatures of invariant NKT cells are shared amongst γδT, innate lymphoid, and Th cells. J. Immunol. 197, 1460–1470 (2016).
Wang, H. & Hogquist, K. A. CCR7 defines a precursor for murine iNKT cells in thymus and periphery. eLife 7, e34793 (2018).
Tuttle, K. D. et al. TCR signal strength controls thymic differentiation of iNKT cell subsets. Nat. Commun. 9, 2650 (2018).
Lee, M. et al. Single-cell RNA sequencing identifies shared differentiation paths of mouse thymic innate T cells. Nat. Commun. 11, 4367 (2020).
Baranek, T., de Amat Herbozo, C., Mallevaey, T. & Paget, C. Deconstructing iNKT cell development at single-cell resolution. Trends Immunol. 43, 503–512 (2022).
Pear, W. S., Tu, L. & Stein, P. L. Lineage choices in the developing thymus: choosing the T and NKT pathways. Curr. Opin. Immunol. 16, 167–173 (2004).
Bezbradica, J. S., Hill, T., Stanic, A. K., Van Kaer, L. & Joyce, S. Commitment toward the natural T (iNKT) cell lineage occurs at the CD4+8+ stage of thymic ontogeny. Proc Natl Acad Sci USA 102, 5114–5119 (2005).
Ligons, D. L. et al. RORγt limits the amount of the cytokine receptor γc through the prosurvival factor Bcl-x(L) in developing thymocytes. Sci. Signal. 11, eaam8939 (2018).
Kurebayashi, S. et al. Retinoid-related orphan receptor γ (RORγ) is essential for lymphoid organogenesis and controls apoptosis during thymopoiesis. Proc. Natl Acad. Sci. USA 97, 10132–10137 (2000).
Sun, Z. et al. Requirement for RORγ in thymocyte survival and lymphoid organ development. Science 288, 2369–2373 (2000).
Guo, J. et al. Regulation of the TCRα repertoire by the survival window of CD4(+)CD8(+) thymocytes. Nat. Immunol. 3, 469–476 (2002).
Bortoluzzi, S. et al. Brief homogeneous TCR signals instruct common iNKT progenitors whose effector diversification is characterized by subsequent cytokine signaling. Immunity 54, 2497–2513 e2499 (2021).
Klibi, J., Amable, L. & Benlagha, K. A focus on natural killer T-cell subset characterization and developmental stages. Immunol. Cell Biol. 98, 358–368 (2020).
Schnell, A., Littman, D. R. & Kuchroo, V. K. T(H)17 cell heterogeneity and its role in tissue inflammation. Nat. Immunol. 24, 19–29 (2023).
Doisne, J. M. et al. iNKT cell development is orchestrated by different branches of TGF-β signaling. J. Exp. Med. 206, 1365–1378 (2009).
Rachitskaya, A. V. et al. Cutting edge: NKT cells constitutively express IL-23 receptor and RORγt and rapidly produce IL-17 upon receptor ligation in an IL-6-independent fashion. J. Immunol. 180, 5167–5171 (2008).
Michel, M. L. et al. Critical role of ROR-γt in a new thymic pathway leading to IL-17-producing invariant NKT cell differentiation. Proc. Natl Acad. Sci. USA 105, 19845–19850 (2008).
Moreira-Teixeira, L. et al. Proinflammatory environment dictates the IL-17-producing capacity of human invariant NKT cells. J. Immunol. 186, 5758–5765 (2011).
Coquet, J. M. et al. Diverse cytokine production by NKT cell subsets and identification of an IL-17-producing CD4-NK1.1- NKT cell population. Proc. Natl Acad. Sci. USA 105, 11287–11292 (2008).
Stritesky, G. L., Yeh, N. & Kaplan, M. H. IL-23 promotes maintenance but not commitment to the Th17 lineage. J. Immunol. 181, 5948–5955 (2008).
Keefe, R., Dave, V., Allman, D., Wiest, D. & Kappes, D. J. Regulation of lineage commitment distinct from positive selection. Science 286, 1149–1153 (1999).
Kappes, D. J. Expanding roles for ThPOK in thymic development. Immunol. Rev. 238, 182–194 (2010).
He, X. et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature 433, 826–833 (2005).
Sun, G. et al. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat. Immunol. 6, 373–381 (2005).
Wang, L. et al. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells. Nat. Immunol. 9, 1122–1130 (2008).
Wang, L. et al. The sequential activity of Gata3 and Thpok is required for the differentiation of CD1d-restricted CD4+ NKT cells. Eur. J. Immunol. 40, 2385–2390 (2010).
Bedel, R. et al. Effective functional maturation of invariant natural killer T cells is constrained by negative selection and T-cell antigen receptor affinity. Proc. Natl Acad. Sci. USA 111, E119–E128 (2014).
Egawa, T. & Littman, D. R. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat. Immunol. 9, 1131–1139 (2008).
Engel, I. et al. Co-receptor choice by Vα14i NKT cells is driven by Th-POK expression rather than avoidance of CD8-mediated negative selection. J. Exp. Med. 207, 1015–1029 (2010).
Luckey, M. A. et al. The transcription factor ThPOK suppresses Runx3 and imposes CD4(+) lineage fate by inducing the SOCS suppressors of cytokine signaling. Nat. Immunol. 15, 638–645 (2014).
Enders, A. et al. ZBTB7B (Th-POK) regulates the development of IL-17-producing CD1d-restricted mouse NKT cells. J. Immunol. 189, 5240–5249 (2012).
Engel, I., Zhao, M., Kappes, D., Taniuchi, I. & Kronenberg, M. The transcription factor Th-POK negatively regulates Th17 differentiation in Vα14i NKT cells. Blood 120, 4524–4532 (2012).
He, X. et al. CD4-CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity 28, 346–358 (2008).
Wildt, K. F. et al. The transcription factor Zbtb7b promotes CD4 expression by antagonizing Runx-mediated activation of the CD4 silencer. J. Immunol. 179, 4405–4414 (2007).
Egawa, T. & Taniuchi, I. Antagonistic interplay between ThPOK and Runx in lineage choice of thymocytes. Blood Cells Mol. Dis. 43, 27–29 (2009).
Liu, X. et al. Runt-related transcription factor 3 is involved in the altered phenotype and function in ThPok-deficient invariant natural killer T cells. Cell. Mol. Immunol. 11, 232–244 (2014).
Levanon, D. & Groner, Y. Structure and regulated expression of mammalian RUNX genes. Oncogene 23, 4211–4219 (2004).
Woolf, E. et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci USA 100, 7731–7736 (2003).
Ohno, S. et al. Runx proteins are involved in regulation of CD122, Ly49 family and IFN-γ expression during NK cell differentiation. Int. Immunol. 20, 71–79 (2008).
Thapa, P. et al. The differentiation of ROR-γt expressing iNKT17 cells is orchestrated by Runx1. Sci. Rep. 7, 7018 (2017).
Lazarevic, V. et al. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORγt. Nat. Immunol. 12, 96–104 (2011).
Yu, J. S. et al. Differentiation of IL-17-producing invariant natural killer T cells requires expression of the transcription factor c-Maf. Front. Immunol. 8, 1399 (2017).
Schraml, B. U. et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature 460, 405–409 (2009).
Thapa, P. et al. NKAP regulates invariant NKT cell proliferation and differentiation into ROR-γt-expressing NKT17 cells. J. Immunol. 196, 4987–4998 (2016).
Drees, C. et al. Roquin paralogs differentially regulate functional NKT cell subsets. J. Immunol. 198, 2747–2759 (2017).
Uddin, M. N. et al. Transcription factor Bcl11b sustains iNKT1 and iNKT2 cell programs, restricts iNKT17 cell program, and governs iNKT cell survival. Proc. Natl Acad. Sci. USA 113, 7608–7613 (2016).
Michel, M. L. et al. Identification of an IL-17-producing NK1.1(neg) iNKT cell population involved in airway neutrophilia. J. Exp. Med. 204, 995–1001 (2007).
Krovi, S. H., Loh, L., Spengler, A., Brunetti, T. & Gapin, L. Current insights in mouse iNKT and MAIT cell development using single cell transcriptomics data. Semin. Immunol. 60, 101658 (2022).
Godfrey, D. I., MacDonald, H. R., Kronenberg, M., Smyth, M. J. & Van Kaer, L. NKT cells: what’s in a name? Nat. Rev. Immunol. 4, 231–237 (2004).
Engel, I. et al. Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nat. Immunol. 17, 728–739 (2016).
Luo, S., Kwon, J., Crossman, A., Park, P. W. & Park, J. H. CD138 expression is a molecular signature but not a developmental requirement for RORγt+ NKT17 cells. JCI Insight 6, e148038 (2021).
Gordy, L. E. et al. IL-15 regulates homeostasis and terminal maturation of NKT cells. J. Immunol. 187, 6335–6345 (2011).
Park, J. Y. et al. In vivo availability of the cytokine IL-7 constrains the survival and homeostasis of peripheral iNKT cells. Cell Rep. 38, 110219 (2022).
Hirota, K. et al. Preferential recruitment of CCR6-expressing Th17 cells to inflamed joints via CCL20 in rheumatoid arthritis and its animal model. J. Exp. Med. 204, 2803–2812 (2007).
Schutyser, E., Struyf, S. & Van Damme, J. The CC chemokine CCL20 and its receptor CCR6. Cytokine Growth Factor Rev. 14, 409–426 (2003).
Dai, H. et al. Syndecan-1 identifies and controls the frequency of IL-17-producing naive natural killer T (NKT17) cells in mice. Eur. J. Immunol. 45, 3045–3051 (2015).
Palaiologou, M., Delladetsima, I. & Tiniakos, D. CD138 (syndecan-1) expression in health and disease. Histol. Histopathol. 29, 177–189 (2014).
Luo, S. et al. The cytokine receptor DR3 identifies and promotes the activation of thymic NKT17 cells. Cell. Mol. Life Sci. 80, 76 (2023).
Yu, Y., Jiang, P., Sun, P., Su, N. & Lin, F. Analysis of therapeutic potential of preclinical models based on DR3/TL1A pathway modulation (review). Exp. Ther. Med. 22, 693 (2021).
Nishikii, H. et al. DR3 signaling modulates the function of Foxp3+ regulatory T cells and the severity of acute graft-versus-host disease. Blood 128, 2846–2858 (2016).
Meylan, F. et al. The TNF-family cytokine TL1A promotes allergic immunopathology through group 2 innate lymphoid cells. Mucosal Immunol. 7, 958–968 (2014).
Terashima, A. et al. A novel subset of mouse NKT cells bearing the IL-17 receptor B responds to IL-25 and contributes to airway hyperreactivity. J. Exp. Med. 205, 2727–2733 (2008).
Watarai, H. et al. Development and function of invariant natural killer T cells producing T(h)2- and T(h)17-cytokines. PLoS Biol. 10, e1001255 (2012).
Pappu, B. P. et al. TL1A-DR3 interaction regulates Th17 cell function and Th17-mediated autoimmune disease. J. Exp. Med. 205, 1049–1062 (2008).
Ranson, T. et al. IL-15 is an essential mediator of peripheral NK-cell homeostasis. Blood 101, 4887–4893 (2003).
Kim, G. Y., Hong, C. & Park, J.-H. Seeing is believing: illuminating the source of in vivo interleukin-7. Immune Netw. 11, 1–10 (2011).
Zhang, H. et al. iNKT subsets differ in their developmental and functional requirements on Foxo1. Proc. Natl Acad. Sci. USA 118, e2105950118 (2021).
Acknowledgements
We apologize to the investigators whose studies could not be discussed and cited in this review because of space limitations. We thank members of the Park laboratory for critically reviewing this manuscript. The figures were created with BioRender.com. This study was supported by the Intramural Research Program of the US National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Funding
Open Access funding provided by the National Institutes of Health (NIH).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liman, N., Park, JH. Markers and makers of NKT17 cells. Exp Mol Med 55, 1090–1098 (2023). https://doi.org/10.1038/s12276-023-01015-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s12276-023-01015-y