Abstract
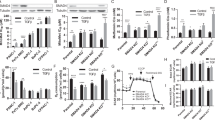
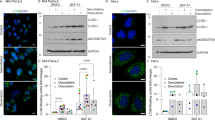
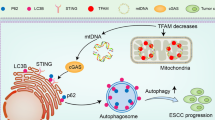
High CD44 expression is associated with enhanced malignant potential in esophageal squamous cell carcinoma (ESCC), among the deadliest of all human carcinomas. Although alterations in autophagy and CD44 expression are associated with poor patient outcomes in various cancer types, the relationship between autophagy and cells with high CD44 expression remains incompletely understood. In transformed oesophageal keratinocytes, CD44Low-CD24High (CD44L) cells give rise to CD44High-CD24-/Low (CD44H) cells via epithelial-mesenchymal transition (EMT) in response to transforming growth factor (TGF)-β. We couple patient samples and xenotransplantation studies with this tractable in vitro system of CD44L to CD44H cell conversion to investigate the functional role of autophagy in generation of cells with high CD44 expression. We report that high expression of the autophagy marker cleaved LC3 expression correlates with poor clinical outcome in ESCC. In ESCC xenograft tumours, pharmacological autophagy inhibition with chloroquine derivatives depletes cells with high CD44 expression while promoting oxidative stress. Autophagic flux impairment during EMT-mediated CD44L to CD44H cell conversion in vitro induces mitochondrial dysfunction, oxidative stress and cell death. During CD44H cell generation, transformed keratinocytes display evidence of mitophagy, including mitochondrial fragmentation, decreased mitochondrial content and mitochondrial translocation of Parkin, essential in mitophagy. RNA interference-mediated Parkin depletion attenuates CD44H cell generation. These data suggest that autophagy facilitates EMT-mediated CD44H generation via modulation of redox homeostasis and Parkin-dependent mitochondrial clearance. This is the first report to implicate mitophagy in regulation of tumour cells with high CD44 expression, representing a potential novel therapeutic avenue in cancers where EMT and CD44H cells have been implicated, including ESCC.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout










Similar content being viewed by others
Abbreviations
- ATG:
-
autophagy-related
- AV(s):
-
autophagic vesicle(s)
- CD44H:
-
CD44High-CD24-/Low
- CD44L:
-
CD44Low-CD24High
- CD44s:
-
standard isoform of CD44
- CD44T:
-
CD44High-CD24High
- CD44v:
-
variant isoforms of CD44
- CQ:
-
Chloroquine
- DAPI:
-
4',6-diamidino-2-phenylindole
- DCF:
-
2′7′-dichlorodihydrofluorescein diacetate
- DPBS:
-
Dulbecco’s Phosphate-buffered Saline
- EMT:
-
epithelial-mesenchymal transition
- EPC2T:
-
EPC2-hTERT-EGFR-p53R175H-CyclinD1
- ESCC:
-
esophageal squamous cell carcinoma
- ESRP:
-
epithelial splicing regulatory protein
- FACS:
-
fluorescence-activated cell sorting
- GAPDH:
-
Glyceraldehyde 3-phosphate dehydrogenase
- HCQ:
-
Hydroxychloroquine
- IHC:
-
Immunohistochemistry
- KSFM:
-
keratinocyte-serum free medium
- LC3:
-
microtubule-associated protein 1 light chain 3
- Mdivi-1:
-
mitochondria division inhibitor-1
- MTCO1:
-
Mitochondrial encoded Cytochrome Oxidase I
- mtDNA:
-
mitochondrial DNA
- O2−:
-
superoxide
- OKF6T:
-
OKF6-hTERT-EGFR-p53R175H
- p-H2A.XSer139:
-
phospho-Histone H2A.X Serine139
- PCR:
-
polymerase chain reaction
- ROS:
-
reactive oxygen species
- RT:
-
Reverse-transcription
- sd:
-
standard deviation
- sem:
-
standard error of the mean
- SOD:
-
superoxide dismutase
- TEM:
-
transmission electron microscopy
- TGF:
-
transforming growth factor
References
Biddle A, Gammon L, Liang X, Costea DE, Mackenzie IC . Phenotypic plasticity determines cancer stem cell therapeutic resistance in oral squamous cell carcinoma. EBioMedicine 2016; 4: 138–145.
Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci USA 2007; 104: 973–978.
Zoller M . CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer 2011; 11: 254–267.
Zhao JS, Li WJ, Ge D, Zhang PJ, Li JJ, Lu CL et al. Tumor initiating cells in esophageal squamous cell carcinomas express high levels of CD44. PLoS One 2011; 6: e21419.
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008; 133: 704–715.
Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A . Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One 2008; 3: e2888.
Kinugasa H, Whelan KA, Tanaka K, Natsuizaka M, Long A, Guo A et al. Mitochondrial SOD2 regulates epithelial-mesenchymal transition and cell populations defined by differential CD44 expression. Oncogene 2015; 34: 5229–5239.
Uchikado Y, Natsugoe S, Okumura H, Setoyama T, Matsumoto M, Ishigami S et al. Slug expression in the E-cadherin preserved tumors is related to prognosis in patients with esophageal squamous cell carcinoma. Clin Cancer Res: An Official Journal of the American Association for Cancer Research 2005; 11: 1174–1180.
Usami Y, Satake S, Nakayama F, Matsumoto M, Ohnuma K, Komori T et al. Snail-associated epithelial-mesenchymal transition promotes oesophageal squamous cell carcinoma motility and progression. J Pathol 2008; 215: 330–339.
Yuen HF, Chan YP, Wong ML, Kwok WK, Chan KK, Lee PY et al. Upregulation of twist in oesophageal squamous cell carcinoma is associated with neoplastic transformation and distant metastasis. J Clin Pathol 2007; 60: 510–514.
Marino G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lopez-Otin C . Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem 2007; 282: 18573–18583.
Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 2003; 112: 1809–1820.
Yue Z, Jin S, Yang C, Levine AJ, Heintz N . Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA 2003; 100: 15077–15082.
Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev 2007; 21: 1621–1635.
Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev 2007; 21: 1367–1381.
Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell 2009; 137: 1062–1075.
Takahashi Y, Hori T, Cooper TK, Liao J, Desai N, Serfass JM et al. Bif-1 haploinsufficiency promotes chromosomal instability and accelerates Myc-driven lymphomagenesis via suppression of mitophagy. Blood 2013; 121: 1622–1632.
Kenific CM, Debnath J . Cellular and metabolic functions for autophagy in cancer cells. Trends Cell Biol 2015; 25: 37–45.
Chen Y, Li X, Guo L, Wu X, He C, Zhang S et al. Combining radiation with autophagy inhibition enhances suppression of tumor growth and angiogenesis in esophageal cancer. Mol Med Rep 2015; 12: 1645–1652.
Yu L, Gu C, Zhong D, Shi L, Kong Y, Zhou Z et al. Induction of autophagy counteracts the anticancer effect of cisplatin in human esophageal cancer cells with acquired drug resistance. Cancer Lett 2014; 355: 34–45.
Enzinger PC, Mayer RJ . Esophageal cancer. N Engl J Med 2003; 349: 2241–2252.
Kleinberg L, Gibson MK, Forastiere AA . Chemoradiotherapy for localized esophageal cancer: regimen selection and molecular mechanisms of radiosensitization. Nat Clin Pract Oncol 2007; 4: 282–294.
Narendra D, Tanaka A, Suen DF, Youle RJ . Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 2008; 183: 795–803.
Amaravadi RK, Winkler JD . Lys05: a new lysosomal autophagy inhibitor. Autophagy 2012; 8: 1383–1384.
McAfee Q, Zhang Z, Samanta A, Levi SM, Ma XH, Piao S et al. Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc Natl Acad Sci USA 2012; 109: 8253–8258.
Ohashi S, Natsuizaka M, Wong GS, Michaylira CZ, Grugan KD, Stairs DB et al. Epidermal growth factor receptor and mutant p53 expand an esophageal cellular subpopulation capable of epithelial-to-mesenchymal transition through ZEB transcription factors. Cancer Res 2010; 70: 4174–4184.
Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J et al. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest 2011; 121: 1064–1074.
Martegani MP, Del Prete F, Gasbarri A, Natali PG, Bartolazzi A . Structural variability of CD44v molecules and reliability of immunodetection of CD44 isoforms using mAbs specific for CD44 variant exon products. Am J Pathol 1999; 154: 291–300.
Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP . ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell 2009; 33: 591–601.
Nozoe T, Kohnoe S, Ezaki T, Kabashima A, Maehara Y . Significance of immunohistochemical over-expression of CD44v6 as an indicator of malignant potential in esophageal squamous cell carcinoma. J Cancer Res Clin Oncol 2004; 130: 334–338.
Shiozaki M, Ishiguro H, Kuwabara Y, Kimura M, Mitsui A, Naganawa Y et al. Expression of CD44v6 is an independent prognostic factor for poor survival in patients with esophageal squamous cell carcinoma. Oncol Lett 2011; 2: 429–434.
Yang H, Liu J, Yu H, Sun P, Hu Y, Zhong J et al. Expression and association of CD44v6 with prognosis in T2-3N0M0 esophageal squamous cell carcinoma. J Thorac Dis 2014; 6: 91–98.
Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016; 12: 1–222.
Cassidy-Stone A, Chipuk JE, Ingerman E, Song C, Yoo C, Kuwana T et al. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell 2008; 14: 193–204.
Cufi S, Vazquez-Martin A, Oliveras-Ferraros C, Martin-Castillo B, Vellon L, Menendez JA . Autophagy positively regulates the CD44(+) CD24(-/low) breast cancer stem-like phenotype. Cell Cycle 2011; 10: 3871–3885.
Lv Q, Wang W, Xue J, Hua F, Mu R, Lin H et al. DEDD interacts with PI3KC3 to activate autophagy and attenuate epithelial-mesenchymal transition in human breast cancer. Cancer Res 2012; 72: 3238–3250.
Li J, Yang B, Zhou Q, Wu Y, Shang D, Guo Y et al. Autophagy promotes hepatocellular carcinoma cell invasion through activation of epithelial-mesenchymal transition. Carcinogenesis 2013; 34: 1343–1351.
Grassi G, Di Caprio G, Santangelo L, Fimia GM, Cozzolino AM, Komatsu M et al. Autophagy regulates hepatocyte identity and epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions promoting Snail degradation. Cell Death Dis 2015; 6: e1880.
Long A, Giroux V, Whelan KA, Hamilton KE, Tetreault MP, Tanaka K et al. WNT10A promotes an invasive and self-renewing phenotype in esophageal squamous cell carcinoma. Carcinogenesis 2015; 36: 598–606.
Ohashi S, Natsuizaka M, Naganuma S, Kagawa S, Kimura S, Itoh H et al. A NOTCH3-mediated squamous cell differentiation program limits expansion of EMT-competent cells that express the ZEB transcription factors. Cancer Res 2011; 71: 6836–6847.
Natsuizaka M, Kinugasa H, Kagawa S, Whelan KA, Naganuma S, Subramanian H et al. IGFBP3 promotes esophageal cancer growth by suppressing oxidative stress in hypoxic tumor microenvironment. Am J Cancer Res 2014; 4: 29–41.
Reznik E, Miller ML, Senbabaoglu Y, Riaz N, Sarungbam J, Tickoo SK et al. Mitochondrial DNA copy number variation across human cancers. Elife 2016; 5: 1–20.
Yu M . Generation, function and diagnostic value of mitochondrial DNA copy number alterations in human cancers. Life Sci 2011; 89: 65–71.
Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci USA 2005; 102: 719–724.
Tseng LM, Yin PH, Chi CW, Hsu CY, Wu CW, Lee LM et al. Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer. Genes Chromosomes Cancer 2006; 45: 629–638.
Archer SL . Mitochondrial dynamics—mitochondrial fission and fusion in human diseases. N Engl J Med 2013; 369: 2236–2251.
Guha M, Srinivasan S, Ruthel G, Kashina AK, Carstens RP, Mendoza A et al. Mitochondrial retrograde signaling induces epithelial-mesenchymal transition and generates breast cancer stem cells. Oncogene 2014; 33: 5238–5250.
Garcia-Prat L, Martinez-Vicente M, Munoz-Canoves P . Autophagy: a decisive process for stemness. Oncotarget 2016; 7: 12286–12288.
Garcia-Prat L, Martinez-Vicente M, Perdiguero E, Ortet L, Rodriguez-Ubreva J, Rebollo E et al. Autophagy maintains stemness by preventing senescence. Nature 2016; 529: 37–42.
Hoshino A, Mita Y, Okawa Y, Ariyoshi M, Iwai-Kanai E, Ueyama T et al. Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nat Commun 2013; 4: 2308.
Rubio N, Coupienne I, Di Valentin E, Heirman I, Grooten J, Piette J et al. Spatiotemporal autophagic degradation of oxidatively damaged organelles after photodynamic stress is amplified by mitochondrial reactive oxygen species. Autophagy 2012; 8: 1312–1324.
Hao CL, Li Y, Yang HX, Luo RZ, Zhang Y, Zhang MF et al. High level of microtubule-associated protein light chain 3 predicts poor prognosis in resectable esophageal squamous cell carcinoma. Int J Clin Exp Pathol 2014; 7: 4213–4221.
Yoshioka A, Miyata H, Doki Y, Yamasaki M, Sohma I, Gotoh K et al. LC3, an autophagosome marker, is highly expressed in gastrointestinal cancers. Int J Oncol 2008; 33: 461–468.
Sabharwal SS, Schumacker PT . Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles’ heel? Nat Rev Cancer 2014; 14: 709–721.
Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res 2011; 17: 654–666.
Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA et al. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol 2000; 20: 1436–1447.
Kalabis J, Wong GS, Vega ME, Natsuizaka M, Robertson ES, Herlyn M et al. Isolation and characterization of mouse and human esophageal epithelial cells in 3D organotypic culture. Nat Protoc 2012; 7: 235–246.
Whelan KA, Merves JF, Giroux V, Tanaka K, Guo A, Chandramouleeswaran PM et al. Autophagy mediates epithelial cytoprotection in eosinophilic oesophagitis. Gut 2016; e-pub ahead of print 16 February 2016 doi:10.1136/gutjnl-2015-310341.
Darling DS, Stearman RP, Qi Y, Qiu MS, Feller JP . Expression of Zfhep/deltaEF1 protein in palate, neural progenitors, and differentiated neurons. Gene Expr Patterns 2003; 3: 709–717.
Schlossman SF . Leucocyte Typing V: White Cell Differentiation Antigens Proceedings of the Fifth International Workshop and Conference; 3−7 November 1993; Boston, USA Oxford University Press: Oxford; New York, 1995.
Kagawa S, Natsuizaka M, Whelan KA, Facompre N, Naganuma S, Ohashi S et al. Cellular senescence checkpoint function determines differential Notch1-dependent oncogenic and tumor-suppressor activities. Oncogene 2015; 34: 2347–2359.
Kong J, Whelan KA, Laczko D, Dang B, Caro Monroig A, Soroush A et al. Autophagy levels are elevated in Barrett’s esophagus and promote cell survival from acid and oxidative stress. Mol Carcinog 2015; 55: 1526–1541.
Kishimoto T . Leucocyte Typing VI: White Cell Differentiation Antigens Proceedings of the Sixth International Workshop and Conference; 10−14 November 1996; Kobe, Japan Garland Pub: New York, 1998.
Tanaka K, Whelan KA, Chandramouleeswaran PM, Kagawa S, Rustgi SL, Noguchi C et al. ALDH2 modulates autophagy flux to regulate acetaldehyde-mediated toxicity thresholds. Am J Cancer Res 2016; 6: 781–796.
Acknowledgements
We appreciate the support the support of members the Nakagawa and Rustgi laboratories for thoughtful discussions and support. We thank Ben Rhodes, Sanders Chang, Andy Guo, Amanda B. Muir, MD and Ashley Lento, PhD for technical support. We acknowledge the Flow Cytometry and Cell Sorting Resource Laboratory, the Electron Microscopy Resource Laboratory, and the Cell and Developmental Biology Microscopy Core at the University of Pennsylvania. This study was supported by the following NIH Grants: P01CA098101 (KAW, KT, PMC, AJK-S, HN, AKR), K26RR032714 (HN), P30ES013508 University of Pennsylvania Center of Excellence in Environmental Toxicology (HN), K01DK103953 (KAW), F32CA174176 (KAW), T32DK007066 (KAW), the American Cancer Society RP-10-033-01-CCE (AKR), NIH/NIDDK P30DK050306 Center of Molecular Studies in Digestive and Liver Diseases, The Molecular Pathology and Imaging, Molecular Biology/Gene Expression, Cell Culture and Mouse Core Facilities. Additional support was provided by the Pennsylvania Department of Health, Pennsylvania CURE Program Grant (HN). KT is a recipient of the Japan Society for the Promotion of Science Postdoctoral Fellowship. His study was supported by the Grant-in-Aid for challenging Exploratory Research, Grant in Aid for Scientific Research B and Grant in Aid for Scientific Research C from the Ministry of Education, Culture, Sports, Science and Technology of Japan (15K15501 and 26293306 to SN; and 15K10108 to YK).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest. Drs Ravi K Amaravadi and Jeffrey D Winkler have licensed Lys05 to Presage biosciences for development. The other authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Whelan, K., Chandramouleeswaran, P., Tanaka, K. et al. Autophagy supports generation of cells with high CD44 expression via modulation of oxidative stress and Parkin-mediated mitochondrial clearance. Oncogene 36, 4843–4858 (2017). https://doi.org/10.1038/onc.2017.102
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.102
This article is cited by
-
Cancer stem cell fate determination: mito-nuclear communication
Cell Communication and Signaling (2023)
-
Redox balance and autophagy regulation in cancer progression and their therapeutic perspective
Medical Oncology (2022)
-
Single cell transcriptomic analysis reveals cellular diversity of murine esophageal epithelium
Nature Communications (2022)
-
IL-17B/IL-17RB signaling cascade contributes to self-renewal and tumorigenesis of cancer stem cells by regulating Beclin-1 ubiquitination
Oncogene (2021)
-
Mitochondrial dynamics in cancer stem cells
Cellular and Molecular Life Sciences (2021)