Abstract
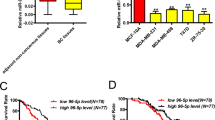
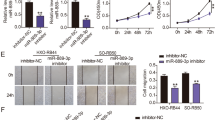
TGF-β has paradoxical effects on cancer cell proliferation, as it suppresses proliferation of normal epithelial and low-invasive cancer cells, but enhances that of high-invasive cancer cells. However, how cancer cells acquire the ability to evade the tumor-suppressing effects of TGF-β, yet still take advantage of its tumor-promoting effects, remains elusive. Here, we identified miR-106b as a molecular switch to determine TGF-β effects on cell proliferation. TGF-β1 enhances the transcription of miR-106b via a promoter independent of its host gene MCM7 by activating c-jun. In high-invasive breast cancer cells, miR-106b is upregulated by TGF-β1 at a much higher level than that in normal or low-invasive cancer cells. Accumulation of miR-106b counterbalances TGF-β growth-inhibiting effects by eliminating activated retinoblastoma (RB) and results in enhanced proliferation. Furthermore, miR-106b mediates TGF-β effects on tumor growth and metastasis in breast cancer xenografts. In addition, miR-106b expression is elevated in higher stage tumors and correlated with tumor progression in breast cancer patients. These findings suggest that high level of miR-106b induced by TGF-β determines the tumor-promoting effects of TGF-β in breast cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Singha PK, Yeh IT, Venkatachalam MA, Saikumar P . Transforming growth factor-beta (TGF-beta)-inducible gene TMEPAI converts TGF-beta from a tumor suppressor to a tumor promoter in breast cancer. Cancer Res 2010; 70: 6377–6383.
Massague J . TGFbeta in Cancer. Cell 2008; 134: 215–230.
Fang Y, Yu S, Braley-Mullen H . TGF-beta promotes proliferation of thyroid epithelial cells in IFN-gamma(-/-) mice by down-regulation of p21 and p27 via AKT pathway. Am J Pathol 2011; 180: 650–660.
Herrera RE, Makela TP, Weinberg RA . TGF beta-induced growth inhibition in primary fibroblasts requires the retinoblastoma protein. Mol Biol Cell 1996; 7: 1335–1342.
Francis SM, Bergsied J, Isaac CE, Coschi CH, Martens AL, Hojilla CV et al. A functional connection between pRB and transforming growth factor beta in growth inhibition and mammary gland development. Mol Cell Biol 2009; 29: 4455–4466.
Furukawa Y, Uenoyama S, Ohta M, Tsunoda A, Griffin JD, Saito M . Transforming growth factor-beta inhibits phosphorylation of the retinoblastoma susceptibility gene product in human monocytic leukemia cell line JOSK-I. J Biol Chem 1992; 267: 17121–17127.
Geng Y, Weinberg RA . Transforming growth factor beta effects on expression of G1 cyclins and cyclin-dependent protein kinases. Proc Natl Acad Sci USA 1993; 90: 10315–10319.
Geng Y, Yu Q, Sicinska E, Das M, Bronson RT, Sicinski P . Deletion of the p27Kip1 gene restores normal development in cyclin D1-deficient mice. Proc Natl Acad Sci USA 2001; 98: 194–199.
Kim TA, Ravitz MJ, Wenner CE . Transforming growth factor-beta regulation of retinoblastoma gene product and E2F transcription factor during cell cycle progression in mouse fibroblasts. J Cell Physiol 1994; 160: 1–9.
Kan T, Sato F, Ito T, Matsumura N, David S, Cheng Y et al. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology 2009; 136: 1689–1700.
Li Y, Tan W, Neo TW, Aung MO, Wasser S, Lim SG et al. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Sci 2009; 100: 1234–1242.
Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 2008; 13: 272–286.
Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM et al. Identification of the miR-106b∼25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal 2010; 3: ra29.
Wang H, Liu J, Zong Y, Xu Y, Deng W, Zhu H et al. miR-106b aberrantly expressed in a double transgenic mouse model for Alzheimer’s disease targets TGF-beta type II receptor. Brain Res 2010; 1357: 166–174.
Smith AL, Iwanaga R, Drasin DJ, Micalizzi DS, Vartuli RL, Tan AC et al. The miR-106b-25 cluster targets Smad7, activates TGF-beta signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene 2012; 31: 5162–5171.
Cai K, Wang Y, Bao X . MiR-106b promotes cell proliferation via targeting RB in laryngeal carcinoma. J Exp Clin Cancer Res 2011; 30: 73.
Kappelmann M, Kuphal S, Meister G, Vardimon L, Bosserhoff AK . MicroRNA miR-125b controls melanoma progression by direct regulation of c-Jun protein expression. Oncogene 2012; 32: 2984–2991.
Zhu F, Choi BY, Ma WY, Zhao Z, Zhang Y, Cho YY et al. COOH-terminal Src kinase-mediated c-Jun phosphorylation promotes c-Jun degradation and inhibits cell transformation. Cancer Res 2006; 66: 5729–5736.
Shaulian E, Karin M . AP-1 as a regulator of cell life and death. Nat Cell Biol 2002; 4: E131–E136.
Behrens A, Jochum W, Sibilia M, Wagner EF . Oncogenic transformation by ras and fos is mediated by c-Jun N-terminal phosphorylation. Oncogene 2000; 19: 2657–2663.
Davies M, Robinson M, Smith E, Huntley S, Prime S, Paterson I . Induction of an epithelial to mesenchymal transition in human immortal and malignant keratinocytes by TGF-beta1 involves MAPK, Smad and AP-1 signalling pathways. J Cell Biochem 2005; 95: 918–931.
Lee MK, Pardoux C, Hall MC, Lee PS, Warburton D, Qing J et al. TGF-beta activates Erk MAP kinase signalling through direct phosphorylation of ShcA. EMBO J 2007; 26: 3957–3967.
Zhang B, Halder SK, Kashikar ND, Cho YJ, Datta A, Gorden DL et al. Antimetastatic role of Smad4 signaling in colorectal cancer. Gastroenterology 2010; 138: 969–980 e961-963.
Burkhart DL, Sage J . Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer 2008; 8: 671–682.
van den Heuvel S, Dyson NJ . Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol 2008; 9: 713–724.
Trimarchi JM, Lees JA . Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol 2002; 3: 11–20.
Acknowledgements
This work was supported by grants from 973 (2010CB912800, 2011CB504203) Projects from Ministry of Science and Technology of China, the Natural Science Foundation of China (81230060, 81261140373, 81272893, 81172524, 81372817, 81272894, 81072178), National S&T Major Special Project on New Drug Innovation of China (No. 2011ZX09102-010-02), Program for New Century Excellent Talents in University(NCET-12-0565), Science Foundation of Guangdong Province (S2012030006287, 2012J2200092), Sun Yat-sen University Training Project (11ykpy28, 11ykzd12) and Translational medicine public platform of Guangdong Province(4202037), Grant KLB09001 from the Key Laboratory of Malignant Tumor Gene Regulation and Target Therapy of Guangdong Higher Education Institutes, Sun-Yat-Sen University,Grant [2013]163 from Key Laboratory of Malignant Tumor Molecular Mechanism and Translational Medicine of Guangzhou Bureau of Science and Information Technology.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Gong, C., Qu, S., Liu, B. et al. MiR-106b expression determines the proliferation paradox of TGF-β in breast cancer cells. Oncogene 34, 84–93 (2015). https://doi.org/10.1038/onc.2013.525
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2013.525
Keywords
This article is cited by
-
Recent developments in targeting breast cancer stem cells (BCSCs): a descriptive review of therapeutic strategies and emerging therapies
Medical Oncology (2024)
-
Integrative analysis of miRNA and mRNA profiles reveals that gga-miR-106-5p inhibits adipogenesis by targeting the KLF15 gene in chickens
Journal of Animal Science and Biotechnology (2022)
-
microRNA-93-5p promotes hepatocellular carcinoma progression via a microRNA-93-5p/MAP3K2/c-Jun positive feedback circuit
Oncogene (2020)
-
miR-106b-5p and miR-17-5p could predict recurrence and progression in breast ductal carcinoma in situ based on the transforming growth factor-beta pathway
Breast Cancer Research and Treatment (2019)
-
The miR-106b-25 cluster mediates breast tumor initiation through activation of NOTCH1 via direct repression of NEDD4L
Oncogene (2018)