Abstract
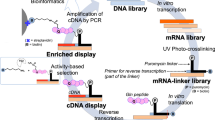
mRNA display is a powerful yet challenging in vitro selection technique that can be used to identify proteins with desired properties from both natural proteome and combinatorial polypeptide libraries. The physical conjugation between a protein and its own RNA presents unique challenges in manipulating the displayed proteins at a low nanomolar scale in an RNase-free environment. The following protocol outlines the generation of cDNA libraries derived from natural organisms as well as the steps required for generation of mRNA-protein fusion molecules, in vitro functional selection and regeneration of the selected cDNA library. The selection procedures for the identification of protease substrates and Ca2+-dependent calmodulin-binding proteins from natural cDNA libraries are presented as examples. The method can be generally applied to the identification of protein sequences with desired properties from various natural proteome libraries. One round of mRNA display–based selection can be accomplished in ∼7 d.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Tateyama, S. et al. Affinity selection of DNA-binding protein complexes using mRNA display. Nucleic Acids. Res. 34, e27 (2006).
Schilling, O., Huesgen, P.F., Barré, O., Auf dem Keller, U. & Overall, C.M. Characterization of the prime and non-prime active site specificities of proteases by proteome-derived peptide libraries and tandem mass spectrometry. Nat. Protoc. 6, 111–120 (2011).
Gevaert, K. et al. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat. Biotechnol. 21, 566–569 (2003).
Brockstedt, E. et al. Identification of apoptosis-associated proteins in a human Burkitt lymphoma cell line. Cleavage of heterogeneous nuclear ribonucleoprotein A1 by caspase 3. J. Biol. Chem. 273, 28057–28064 (1998).
Fields, S. & Song, O. A novel genetic system to detect protein-protein interactions. Nature 340, 245–246 (1989).
Luban, J. & Goff, S.P. The yeast two-hybrid system for studying protein-protein interactions. Curr. Opin. Biotechnol. 6, 59–64 (1995).
Miller, J. & Stagljar, I. Using the yeast two-hybrid system to identify interacting proteins. Methods Mol. Biol. 261, 247–262 (2004).
Lin, H. & Cornish, V.W. Screening and selection methods for large-scale analysis of protein function. Angew. Chem. Int. Ed. Engl. 41, 4402–4425 (2002).
Smith, G.P. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228, 1315–1317 (1985).
Parmley, S.F. & Smith, G.P. Antibody-selectable filamentous fd phage vectors: affinity purification of target genes. Gene 73, 305–318 (1988).
Smith, G.P. & Scott, J.K. Libraries of peptides and proteins displayed on filamentous phage. Meth. Enzymol. 217, 228–257 (1993).
Conrad, U. & Scheller, J. Considerations on antibody-phage display methodology. Comb. Chem. High Throughput Screen 8, 117–126 (2005).
Weaver-Feldhaus, J.M., Miller, K.D., Feldhaus, M.J. & Siegel, R.W. Directed evolution for the development of conformation-specific affinity reagents using yeast display. Protein Eng. Des. Sel. 18, 527–536 (2005).
Fukuda, N. et al. High-efficiency recovery of target cells using improved yeast display system for detection of protein-protein interactions. Appl. Microbiol. Biotechnol. 76, 151–158 (2007).
Jung, S.T., Jeong, K.J., Iverson, B.L. & Georgiou, G. Binding and enrichment of Escherichia coli spheroplasts expressing inner membrane tethered scFv antibodies on surface immobilized antigens. Biotechnol. Bioeng. 98, 39–47 (2007).
Mattheakis, L.C., Bhatt, R.R. & Dower, W.J. An in vitro polysome display system for identifying ligands from very large peptide libraries. Proc. Natl. Acad. Sci. USA 91, 9022–9026 (1994).
Hanes, J. & Plückthun, A. In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl. Acad. Sci. USA 94, 4937–4942 (1997).
Brückner, A., Polge, C., Lentze, N., Auerbach, D. & Schlattner, U. Yeast two-hybrid, a powerful tool for systems biology. Int. J. Mol. Sci. 10, 2763–2788 (2009).
Huang, H., Jedynak, B.M. & Bader, J.S. Where have all the interactions gone? Estimating the coverage of two-hybrid protein interaction maps. PLoS Comput. Biol. 3, e214 (2007).
Rhyner, C. et al. Cloning allergens via phage display. Methods 32, 212–218 (2004).
Adey, N.B. & Kay, B.K. Identification of calmodulin-binding peptide consensus sequences from a phage-displayed random peptide library. Gene 169, 133–134 (1996).
He, M. & Taussig, M.J. Eukaryotic ribosome display with in situ DNA recovery. Nat. Methods 4, 281–288 (2007).
Zahnd, C., Amstutz, P. & Plückthun, A. Ribosome display: selecting and evolving proteins in vitro that specifically bind to a target. Nat. Methods 4, 269–279 (2007).
Ju, W. et al. Proteome-wide identification of family member-specific natural substrate repertoire of caspases. Proc. Natl. Acad. Sci. USA 104, 14294–14299 (2007).
Shen, X. et al. Scanning the human proteome for calmodulin-binding proteins. Proc. Natl. Acad. Sci. USA 102, 5969–5974 (2005).
Shen, X. et al. Ca(2+)/Calmodulin-binding proteins from the C. elegans proteome. Cell Calcium 43, 444–456 (2008).
Huang, B. & Liu, R. Comparison of mRNA-display-based selections using synthetic peptide and natural protein libraries. Biochemistry 46, 10102–10112 (2007).
Valencia, C.A., Cotten, S.W., Dong, B. & Liu, R. mRNA-display-based selections for proteins with desired functions: a protease-substrate case study. Biotechnol. Prog. 24, 561–569 (2008).
Roberts, R.W. & Szostak, J.W. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl. Acad. Sci. USA 94, 12297–12302 (1997).
Liu, R., Barrick, J.E., Szostak, J.W. & Roberts, R.W. Optimized synthesis of RNA-protein fusions for in vitro protein selection. Meth. Enzymol. 318, 268–293 (2000).
Nemoto, N., Miyamoto-Sato, E., Husimi, Y. & Yanagawa, H. In vitro virus: bonding of mRNA bearing puromycin at the 3′-terminal end to the C-terminal end of its encoded protein on the ribosome in vitro. FEBS Lett. 414, 405–408 (1997).
Hammond, P.W., Alpin, J., Rise, C.E., Wright, M. & Kreider, B.L. In vitro selection and characterization of Bcl-X(L)-binding proteins from a mix of tissue-specific mRNA display libraries. J. Biol. Chem. 276, 20898–20906 (2001).
Cujec, T.P., Medeiros, P.F., Hammond, P., Rise, C. & Kreider, B.L. Selection of v-abl tyrosine kinase substrate sequences from randomized peptide and cellular proteomic libraries using mRNA display. Chem. Biol. 9, 253–264 (2002).
McPherson, M., Yang, Y., Hammond, P.W. & Kreider, B.L. Drug receptor identification from multiple tissues using cellular-derived mRNA display libraries. Chem. Biol. 9, 691–698 (2002).
Horisawa, K. et al. In vitro selection of Jun-associated proteins using mRNA display. Nucleic Acids. Res. 32, e169 (2004).
Fukuda, I. et al. In vitro evolution of single-chain antibodies using mRNA display. Nucleic Acids. Res. 34, e127 (2006).
Josephson, K., Hartman, M.C.T. & Szostak, J.W. Ribosomal synthesis of unnatural peptides. J. Am. Chem. Soc. 127, 11727–11735 (2005).
Seelig, B. & Szostak, J.W. Selection and evolution of enzymes from a partially randomized non-catalytic scaffold. Nature 448, 828–831 (2007).
Wilson, D.S., Keefe, A.D. & Szostak, J.W. The use of mRNA display to select high-affinity protein-binding peptides. Proc. Natl. Acad. Sci. USA 98, 3750–3755 (2001).
Xu, L. et al. Directed evolution of high-affinity antibody mimics using mRNA display. Chem. Biol. 9, 933–942 (2002).
Olson, C.A., Liao, H., Sun, R. & Roberts, R.W. mRNA display selection of a high-affinity, modification-specific phospho-IkappaBalpha-binding fibronectin. ACS Chem. Biol. 3, 480–485 (2008).
Liao, H. et al. mRNA display design of fibronectin-based intrabodies that detect and inhibit severe acute respiratory syndrome coronavirus nucleocapsid protein. J. Biol. Chem. 284, 17512–17520 (2009).
Kosugi, S. et al. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J. Biol. Chem. 284, 478–485 (2009).
Austin, R.J., Ja, W.W. & Roberts, R.W. Evolution of class-specific peptides targeting a hot spot of the Galphas subunit. J. Mol. Biol. 377, 1406–1418 (2008).
Ambion. Retic Lysate IVT Instruction Manual. At 〈http://www.ambion.com/catalog/ProdGrp.html?fkApp=11&fkProdGrp=103〉.
Cho, G., Keefe, A.D., Liu, R., Wilson, D.S. & Szostak, J.W. Constructing high complexity synthetic libraries of long ORFs using in vitro selection. J. Mol. Biol. 297, 309–319 (2000).
Kurz, M., Gu, K. & Lohse, P.A. Psoralen photo-crosslinked mRNA-puromycin conjugates: a novel template for the rapid and facile preparation of mRNA-protein fusions. Nucleic Acids. Res. 28, e83 (2000).
Leemhuis, H., Stein, V., Griffiths, A.D. & Hollfelder, F. New genotype-phenotype linkages for directed evolution of functional proteins. Curr. Opin. Struct. Biol. 15, 472–478 (2005).
Tabuchi, I. et al. An efficient ligation method in the making of an in vitro virus for in vitro protein evolution. Biol. Proced Online 4, 49–54 (2002).
Metzker, M.L. Sequencing technologies—the next generation. Nat. Rev. Genet. 11, 31–46 (2010).
Acknowledgements
This work was supported by American Cancer Society Research Scholar Grant RSG-06-073 (to R.L.) and National Institutes of Health Grants NS047650, CA119343 and DA025702 (to R.L.).
Author information
Authors and Affiliations
Contributions
R.L. designed the experiments and supervised the projects. S.W.C., J.Z., C.A.V. and R.L. conducted the experiments and analyzed the data. S.W.C., J.Z., C.A.V. and R.L. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Cotten, S., Zou, J., Valencia, C. et al. Selection of proteins with desired properties from natural proteome libraries using mRNA display. Nat Protoc 6, 1163–1182 (2011). https://doi.org/10.1038/nprot.2011.354
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2011.354
This article is cited by
-
LOVTRAP: an optogenetic system for photoinduced protein dissociation
Nature Methods (2016)
-
Liposome display for in vitro selection and evolution of membrane proteins
Nature Protocols (2014)
-
Creation of recombinant antigen-binding molecules derived from hybridomas secreting specific antibodies
Nature Protocols (2013)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.