Abstract
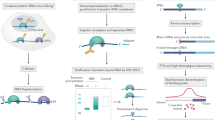
We describe a protocol in which dimethyl sulfate (DMS) modification of the base-pairing faces of unpaired adenosine and cytidine nucleotides is used for structural analysis of RNAs and RNA–protein complexes (RNPs). The protocol is optimized for RNAs of small to moderate size (≤500 nt). The RNA or RNP is first exposed to DMS under conditions that promote formation of the folded structure or complex, as well as 'control' conditions that do not allow folding or complex formation. The positions and extents of modification are then determined by primer extension, polyacrylamide gel electrophoresis and quantitative analysis. From changes in the extent of modification upon folding or protein binding (appearance of a 'footprint'), it is possible to detect local changes in the secondary and tertiary structure of RNA, as well as the formation of RNA–protein contacts. This protocol takes 1.5–3 d to complete, depending on the type of analysis used.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Golden, B.L. Preparation and crystallization of RNA. Methods Mol. Biol. 363, 239–257 (2007).
Wu, H., Finger, L.D. & Feigon, J. Structure determination of protein/RNA complexes by NMR. Methods Enzymol. 394, 525–545 (2005).
Myong, S., Stevens, B.C. & Ha, T. Bridging conformational dynamics and function using single-molecule spectroscopy. Structure 14, 633–643 (2006).
Lipfert, J. & Doniach, S. Small-angle X-ray scattering from RNA, proteins, and protein complexes. Annu. Rev. Biophys. Biomol. Struct. 36, 307–327 (2007).
Kim, N.K., Murali, A. & DeRose, V.J. A distance ruler for RNA using EPR and site-directed spin labeling. Chem. Biol. 11, 939–948 (2004).
Das, R., Travers, K.J., Bai, Y. & Herschlag, D. Determining the Mg2+ stoichiometry for folding an RNA metal ion core. J. Am. Chem. Soc. 127, 8272–8273 (2005).
Su, L.J., Brenowitz, M. & Pyle, A.M. An alternative route for the folding of large RNAs: apparent two-state folding by a group II intron ribozyme. J. Mol. Biol. 334, 639–652 (2003).
Lilley, D.M. Analysis of global conformational transitions in ribozymes. Methods Mol. Biol. 252, 77–108 (2004).
Tullius, T.D. & Greenbaum, J.A. Mapping nucleic acid structure by hydroxyl radical cleavage. Curr. Opin. Chem. Biol. 9, 127–134 (2005).
Brenowitz, M., Chance, M.R., Dhavan, G. & Takamoto, K. Probing the structural dynamics of nucleic acids by quantitative time-resolved and equilibrium hydroxyl radical 'footprinting'. Curr. Opin. Struct. Biol. 12, 648–653 (2002).
Shcherbakova, I., Mitra, S., Beer, R.H. & Brenowitz, M. Fast Fenton footprinting: a laboratory-based method for the time-resolved analysis of DNA, RNA and proteins. Nucleic Acids Res. 34, e48 (2006).
Culver, G.M. & Noller, H.F. Directed hydroxyl radical probing of RNA from iron(II) tethered to proteins in ribonucleoprotein complexes. Methods Enzymol. 318, 461–475 (2000).
Black, D.L. & Pinto, A.L. U5 small nuclear ribonucleoprotein: RNA structure analysis and ATP-dependent interaction with U4/U6. Mol. Cell. Biol. 9, 3350–3359 (1989).
Zarrinkar, P.P. & Williamson, J.R. Kinetic intermediates in RNA folding. Science 265, 918–924 (1994).
Ryder, S.P., Ortoleva-Donnelly, L., Kosek, A.B. & Strobel, S.A. Chemical probing of RNA by nucleotide analog interference mapping. Methods Enzymol. 317, 92–109 (2000).
Soukup, G.A. & Breaker, R.R. Relationship between internucleotide linkage geometry and the stability of RNA. RNA 5, 1308–1325 (1999).
Merino, E.J., Wilkinson, K.A., Coughlan, J.L. & Weeks, K.M. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 127, 4223–4231 (2005).
Wilkinson, K.A., Merino, E.J. & Weeks, K.M. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 1, 1610–1616 (2006).
Mortimer, S.A. & Weeks, K.M. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 129, 4144–4145 (2007).
Russell, R. & Herschlag, D. New pathways in folding of the Tetrahymena group I RNA enzyme. J. Mol. Biol. 291, 1155–1167 (1999).
Peattie, D.A. & Gilbert, W. Chemical probes for higher-order structure in RNA. Proc. Natl. Acad. Sci. USA 77, 4679–4682 (1980).
Maxam, A.M. & Gilbert, W. A new method for sequencing DNA. Proc. Natl. Acad. Sci. USA 74, 560–564 (1977).
Peattie, D.A. Direct chemical method for sequencing RNA. Proc. Natl. Acad. Sci. USA 76, 1760–1764 (1979).
Gilbert, W., Maxam, A.M. & Mirzabekov, A. in Control of Ribosome Synthesis (eds. Kjeldgaard, N.O. & Malaloe, O.) 139–148 (Academic, New York, 1976).
Johnsrud, L. Contacts between Escherichia coli RNA polymerase and a lac operon promoter. Proc. Natl. Acad. Sci. USA 75, 5314–5318 (1978).
Siebenlist, U. & Gilbert, W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc. Natl. Acad. Sci. USA 77, 122–126 (1980).
Lawley, P.D. & Brookes, P. Further studies on the alkylation of nucleic acids and their constituent nucleotides. Biochem. J. 89, 127–138 (1963).
Inoue, T. & Cech, T. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc. Natl. Acad. Sci. USA 82, 648–652 (1985).
Lempereur, L. et al. Conformation of yeast 18S rRNA. Direct chemical probing of the 5′ domain in ribosomal subunits and in deproteinized RNA by reverse transcriptase mapping of dimethyl sulfate-accessible sites. Nucleic Acids Res. 13, 8339–8357 (1985).
Moazed, D., Stern, S. & Noller, H.F. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J. Mol. Biol. 187, 399–416 (1986).
Stern, S., Wilson, R.C. & Noller, H.F. Localization of the binding site for protein S4 on 16 S ribosomal RNA by chemical and enzymatic probing and primer extension. J. Mol. Biol. 192, 101–110 (1986).
Moazed, D. & Noller, H.F. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell 47, 985–994 (1986).
Egebjerg, J., Leffers, H., Christensen, A., Andersen, H. & Garrett, R.A. Structure and accessibility of domain I of Escherichia coli 23 S RNA in free RNA, in the L24-RNA complex and in 50 S subunits. Implications for ribosomal assembly. J. Mol. Biol. 196, 125–136 (1987).
Baudin, F. et al. Higher-order structure of domain III in Escherichia coli 16S ribosomal RNA, 30S subunit and 70S ribosome. Biochimie 69, 1081–1096 (1987).
Mougel, M., Philippe, C., Ebel, J.P., Ehresmann, B. & Ehresmann, C. The E. coli 16S rRNA binding site of ribosomal protein S15: higher-order structure in the absence and in the presence of the protein. Nucleic Acids Res. 16, 2825–2839 (1988).
Svensson, P., Changchien, L.M., Craven, G.R. & Noller, H.F. Interaction of ribosomal proteins, S6, S8, S15 and S18 with the central domain of 16 S ribosomal RNA. J. Mol. Biol. 200, 301–308 (1988).
Stern, S., Changchien, L.M., Craven, G.R. & Noller, H.F. Interaction of proteins S16, S17 and S20 with 16 S ribosomal RNA. J. Mol. Biol. 200, 291–299 (1988).
Powers, T., Changchien, L.M., Craven, G.R. & Noller, H.F. Probing the assembly of the 3′ major domain of 16 S ribosomal RNA. Quaternary interactions involving ribosomal proteins S7, S9 and S19. J. Mol. Biol. 200, 309–319 (1988).
Powers, T., Stern, S., Changchien, L.M. & Noller, H.F. Probing the assembly of the 3′ major domain of 16 S rRNA. Interactions involving ribosomal proteins S2, S3, S10, S13 and S14. J. Mol. Biol. 201, 697–716 (1988).
Moazed, D., Robertson, J.M. & Noller, H.F. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature 334, 362–364 (1988).
Moazed, D. & Noller, H.F. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell 57, 585–597 (1989).
Moazed, D. & Noller, H.F. Intermediate states in the movement of transfer RNA in the ribosome. Nature 342, 142–148 (1989).
Stern, S., Powers, T., Changchien, L.M. & Noller, H.F. RNA–protein interactions in 30S ribosomal subunits: folding and function of 16S rRNA. Science 244, 783–790 (1989).
Moazed, D. & Noller, H.F. Sites of interaction of the CCA end of peptidyl-tRNA with 23S rRNA. Proc. Natl. Acad. Sci. USA 88, 3725–3728 (1991).
Banerjee, A.R., Jaeger, J.A. & Turner, D.H. Thermal unfolding of a group I ribozyme: the low-temperature transition is primarily disruption of tertiary structure. Biochemistry 32, 153–163 (1993).
Murphy, F.L. & Cech, T.R. An independently folding domain of RNA tertiary structure within the Tetrahymena ribozyme. Biochemistry 32, 5291–5300 (1993).
Mohr, G., Caprara, M.G., Guo, Q. & Lambowitz, A.M. A tyrosyl-tRNA synthetase can function similarly to an RNA structure in the Tetrahymena ribozyme. Nature 370, 147–150 (1994).
Emerick, V.L. & Woodson, S.A. Fingerprinting the folding of a group I precursor RNA. Proc. Natl. Acad. Sci. USA 91, 9675–9679 (1994).
Doherty, E.A. & Doudna, J.A. The P4–P6 domain directs higher order folding of the Tetrahymena ribozyme core. Biochemistry 36, 3159–3169 (1997).
Caprara, M.G., Myers, C.A. & Lambowitz, A.M. Interaction of the Neurospora crassa mitochondrial tyrosyl-tRNA synthetase (CYT-18 protein) with the group I intron P4–P6 domain. Thermodynamic analysis and the role of metal ions. J. Mol. Biol. 308, 165–190 (2001).
Russell, R. et al. Exploring the folding landscape of a structured RNA. Proc. Natl. Acad. Sci. USA 99, 155–160 (2002).
Mohr, S., Stryker, J.M. & Lambowitz, A.M. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell 109, 769–779 (2002).
Russell, R. et al. The paradoxical behavior of a highly structured misfolded intermediate in RNA folding. J. Mol. Biol. 363, 531–544 (2006).
Russell, R., Tijerina, P., Chadee, A.B. & Bhaskaran, H. Deletion of the P5abc peripheral element accelerates early and late folding steps of the Tetrahymena group I ribozyme. Biochemistry 46, 4951–4961 (2007).
Costa, M., Christian, E.L. & Michel, F. Differential chemical probing of a group II self-splicing intron identifies bases involved in tertiary interactions and supports an alternative secondary structure model of domain V. RNA 4, 1055–1068 (1998).
Konforti, B.B., Liu, Q. & Pyle, A.M. A map of the binding site for catalytic domain 5 in the core of a group II intron ribozyme. EMBO J. 17, 7105–7117 (1998).
Tranguch, A.J., Kindelberger, D.W., Rohlman, C.E., Lee, J.Y. & Engelke, D.R. Structure-sensitive RNA footprinting of yeast nuclear ribonuclease P. Biochemistry 33, 1778–1787 (1994).
Kumar, P.K., Taira, K. & Nishikawa, S. Chemical probing studies of variants of the genomic hepatitis delta virus ribozyme by primer extension analysis. Biochemistry 33, 583–592 (1994).
Butcher, S.E. & Burke, J.M. Structure-mapping of the hairpin ribozyme. Magnesium-dependent folding and evidence for tertiary interactions within the ribozyme–substrate complex. J. Mol. Biol. 244, 52–63 (1994).
Keiper, S., Bebenroth, D., Seelig, B., Westhof, E. & Jaschke, A. Architecture of a Diels-Alderase ribozyme with a preformed catalytic pocket. Chem. Biol. 11, 1217–1227 (2004).
Wang, Y.H., Sczekan, S.R. & Theil, E.C. Structure of the 5′ untranslated regulatory region of ferritin mRNA studied in solution. Nucleic Acids Res. 18, 4463–4468 (1990).
Rettberg, C.C., Prere, M.F., Gesteland, R.F., Atkins, J.F. & Fayet, O. A three-way junction and constituent stem-loops as the stimulator for programmed -1 frameshifting in bacterial insertion sequence IS911. J. Mol. Biol. 286, 1365–1378 (1999).
Fernandez-Miragall, O. & Martinez-Salas, E. Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA 9, 1333–1344 (2003).
Ruschak, A.M. et al. Secondary structure models of the 3′ untranslated regions of diverse R2 RNAs. RNA 10, 978–987 (2004).
Rose, M.A. & Weeks, K.M. Visualizing induced fit in early assembly of the human signal recognition particle. Nat. Struct. Biol. 8, 515–520 (2001).
Menichelli, E., Isel, C., Oubridge, C. & Nagai, K. Protein-induced conformational changes of RNA during the assembly of human signal recognition particle. J. Mol. Biol. 367, 187–203 (2007).
Krol, A. et al. Solution structure of human U1 snRNA. Derivation of a possible three-dimensional model. Nucleic Acids Res. 18, 3803–3811 (1990).
Hartmuth, K., Raker, V.A., Huber, J., Branlant, C. & Luhrmann, R. An unusual chemical reactivity of Sm site adenosines strongly correlates with proper assembly of core U snRNP particles. J. Mol. Biol. 285, 133–147 (1999).
Ast, G., Pavelitz, T. & Weiner, A.M. Sequences upstream of the branch site are required to form helix II between U2 and U6 snRNA in a trans-splicing reaction. Nucleic Acids Res. 29, 1741–1749 (2001).
Brunel, C., Romby, P., Westhof, E., Ehresmann, C. & Ehresmann, B. Three-dimensional model of Escherichia coli ribosomal 5 S RNA as deduced from structure probing in solution and computer modeling. J. Mol. Biol. 221, 293–308 (1991).
Mathews, D.H. et al. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA 101, 7287–7292 (2004).
Lavery, R. & Pullman, A. A new theoretical index of biochemical reactivity combining steric and electrostatic factors. An application to yeast tRNAPhe. Biophys. Chem. 19, 171–181 (1984).
Muth, G.W., Ortoleva-Donnelly, L. & Strobel, S.A. A single adenosine with a neutral pKa in the ribosomal peptidyl transferase center. Science 289, 947–950 (2000).
Bayfield, M.A., Dahlberg, A.E., Schulmeister, U., Dorner, S. & Barta, A. A conformational change in the ribosomal peptidyl transferase center upon active/inactive transition. Proc. Natl. Acad. Sci. USA 98, 10096–10101 (2001).
Muth, G.W., Chen, L., Kosek, A.B. & Strobel, S.A. pH-dependent conformational flexibility within the ribosomal peptidyl transferase center. RNA 7, 1403–1415 (2001).
Altuvia, S., Kornitzer, D., Teff, D. & Oppenheim, A.B. Alternative mRNA structures of the cIII gene of bacteriophage lambda determine the rate of its translation initiation. J. Mol. Biol. 210, 265–280 (1989).
Matsumura, S., Ikawa, Y. & Inoue, T. Biochemical characterization of the kink-turn RNA motif. Nucleic Acids Res. 31, 5544–5551 (2003).
Egebjerg, J., Christiansen, J., Brown, R.S., Larsen, N. & Garrett, R.A. Protein L18 binds primarily at the junctions of helix II and internal loops A and B in Escherichia coli 5 S RNA. Implications for 5 S RNA structure. J. Mol. Biol. 206, 651–668 (1989).
Moazed, D., Samaha, R.R., Gualerzi, C. & Noller, H.F. Specific protection of 16 S rRNA by translational initiation factors. J. Mol. Biol. 248, 207–210 (1995).
Merryman, C., Moazed, D., McWhirter, J. & Noller, H.F. Nucleotides in 16S rRNA protected by the association of 30S and 50S ribosomal subunits. J. Mol. Biol. 285, 97–105 (1999).
Dahlquist, K.D. & Puglisi, J.D. Interaction of translation initiation factor IF1 with the E. coli ribosomal A site. J. Mol. Biol. 299, 1–15 (2000).
Green, C.D., Long, K.S., Shi, H. & Wolin, S.L. Binding of the 60-kDa Ro autoantigen to Y RNAs: evidence for recognition in the major groove of a conserved helix. RNA 4, 750–765 (1998).
Argaman, L. & Altuvia, S. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Mol. Biol. 300, 1101–1112 (2000).
Moazed, D. & Noller, H.F. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327, 389–394 (1987).
Woodcock, J., Moazed, D., Cannon, M., Davies, J. & Noller, H.F. Interaction of antibiotics with A- and P-site-specific bases in 16S ribosomal RNA. EMBO J. 10, 3099–3103 (1991).
Spickler, C., Brunelle, M.N. & Brakier-Gingras, L. Streptomycin binds to the decoding center of 16 S ribosomal RNA. J. Mol. Biol. 273, 586–599 (1997).
Purohit, P. & Stern, S. Interactions of a small RNA with antibiotic and RNA ligands of the 30S subunit. Nature 370, 659–662 (1994).
Moazed, D., Van Stolk, B.J., Douthwaite, S. & Noller, H.F. Interconversion of active and inactive 30 S ribosomal subunits is accompanied by a conformational change in the decoding region of 16 S rRNA. J. Mol. Biol. 191, 483–493 (1986).
Powers, T., Daubresse, G. & Noller, H.F. Dynamics of in vitro assembly of 16 S rRNA into 30 S ribosomal subunits. J. Mol. Biol. 232, 362–374 (1993).
Powers, T. & Noller, H.F. A temperature-dependent conformational rearrangement in the ribosomal protein S4.16 S rRNA complex. J. Biol. Chem. 270, 1238–1242 (1995).
Karginov, F.V. & Uhlenbeck, O.C. Interaction of Escherichia coli DbpA with 23S rRNA in different functional states of the enzyme. Nucleic Acids Res. 32, 3028–3032 (2004).
Hennelly, S.P. et al. A time-resolved investigation of ribosomal subunit association. J. Mol. Biol. 346, 1243–1258 (2005).
Fabbretti, A. et al. The real-time path of translation factor IF3 onto and off the ribosome. Mol. Cell 25, 285–296 (2007).
Herr, W., Chapman, N.M. & Noller, H.F. Mechanism of ribosomal subunit association: discrimination of specific sites in 16 S RNA essential for association activity. J. Mol. Biol. 130, 433–449 (1979).
Peattie, D.A. & Herr, W. Chemical probing of the tRNA–ribosome complex. Proc. Natl. Acad. Sci. USA 78, 2273–2277 (1981).
von Ahsen, U. & Noller, H.F. Identification of bases in 16S rRNA essential for tRNA binding at the 30S ribosomal P site. Science 267, 234–237 (1995).
Pan, J. & Woodson, S.A. Folding intermediates of a self-splicing RNA: mispairing of the catalytic core. J. Mol. Biol. 280, 597–609 (1998).
Zaug, A.J. & Cech, T.R. Analysis of the structure of Tetrahymena nuclear RNAs in vivo: telomerase RNA, the self-splicing rRNA intron, and U2 snRNA. RNA 1, 363–374 (1995).
Charpentier, B., Stutz, F. & Rosbash, M. A dynamic in vivo view of the HIV-I Rev-RRE interaction. J. Mol. Biol. 266, 950–962 (1997).
Iseni, F., Baudin, F., Blondel, D. & Ruigrok, R.W. Structure of the RNA inside the vesicular stomatitis virus nucleocapsid. RNA 6, 270–281 (2000).
Bricker, A.L. & Belasco, J.G. Importance of a 5′ stem-loop for longevity of papA mRNA in Escherichia coli. J. Bacteriol. 181, 3587–3590 (1999).
Doktycz, M.J., Larimer, F.W., Pastrnak, M. & Stevens, A. Comparative analyses of the secondary structures of synthetic and intracellular yeast MFA2 mRNAs. Proc. Natl. Acad. Sci. USA 95, 14614–14621 (1998).
Luo, D., Condon, C., Grunberg-Manago, M. & Putzer, H. In vitro and in vivo secondary structure probing of the thrS leader in Bacillus subtilis. Nucleic Acids Res. 26, 5379–5387 (1998).
Waldsich, C., Grossberger, R. & Schroeder, R. RNA chaperone StpA loosens interactions of the tertiary structure in the td group I intron in vivo. Genes Dev. 16, 2300–2312 (2002).
Disney, M.D., Haidaris, C.G. & Turner, D.H. Uptake and antifungal activity of oligonucleotides in Candida albicans. Proc. Natl. Acad. Sci. USA 100, 1530–1534 (2003).
Wells, S.E., Hughes, J.M., Igel, A.H. & Ares, M. Jr. Use of dimethyl sulfate to probe RNA structure in vivo. Methods Enzymol. 318, 479–493 (2000).
Ehresmann, C. et al. Probing the structure of RNAs in solution. Nucleic Acids Res. 15, 9109–9128 (1987).
Christiansen, J. & Garrett, R. Enzymatic and chemical probing of ribosomal RNA–protein interactions. Methods Enzymol. 164, 456–468 (1988).
Stern, S., Moazed, D. & Noller, H.F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 164, 481–489 (1988).
Merryman, C. & Noller, H.F. in RNA:Protein Interactions, a Practical Approach (ed. Smith, C.W.J.) 237–253 (Oxford University Press, Oxford, 1998).
Brunel, C. & Romby, P. Probing RNA structure and RNA–ligand complexes with chemical probes. Methods Enzymol. 318, 3–21 (2000).
Sanger, F., Nicklen, S. & Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74, 5463–5467 (1977).
Cate, J.H. et al. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science 273, 1678–1685 (1996).
Nissen, P., Ippolito, J.A., Ban, N., Moore, P.B. & Steitz, T.A. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl. Acad. Sci. USA 98, 4899–4903 (2001).
Balasubramanian, B., Pogozelski, W.K. & Tullius, T.D. DNA strand breaking by the hydroxyl radical is governed by the accessible surface areas of the hydrogen atoms of the DNA backbone. Proc. Natl. Acad. Sci. USA 95, 9738–9743 (1998).
Latham, J.A. & Cech, T.R. Defining the inside and outside of a catalytic RNA molecule. Science 245, 276–282 (1989).
King, P.A., Jamison, E., Strahs, D., Anderson, V.E. & Brenowitz, M. 'Footprinting' proteins on DNA with peroxonitrous acid. Nucleic Acids Res. 21, 2473–2478 (1993).
Chaulk, S.G. & MacMillan, A.M. Characterization of the Tetrahymena ribozyme folding pathway using the kinetic footprinting reagent peroxynitrous acid. Biochemistry 39, 2–8 (2000).
Kent, O., Chaulk, S.G. & MacMillan, A.M. Kinetic analysis of the M1 RNA folding pathway. J. Mol. Biol. 304, 699–705 (2000).
Kwok, L.W. et al. Concordant exploration of the kinetics of RNA folding from global and local perspectives. J. Mol. Biol. 355, 282–293 (2006).
Lindahl, T., Adams, A. & Fresco, J.R. Renaturation of transfer ribonucleic acids through site binding of magnesium. Proc. Natl. Acad. Sci. USA 55, 941–948 (1966).
Weidner, H. & Crothers, D.M. Pathway-dependent refolding of E. coli 5S RNA. Nucleic Acids Res. 4, 3401–3414 (1977).
Sugimoto, N., Kierzek, R. & Turner, D.H. Kinetics for reaction of a circularized intervening sequence with CU, UCU, CUCU, and CUCUCU: mechanistic implications from the dependence on temperature and on oligomer and Mg2+ concentrations. Biochemistry 27, 6384–6392 (1988).
Walstrum, S.A. & Uhlenbeck, O.C. The self-splicing RNA of Tetrahymena is trapped in a less active conformation by gel purification. Biochemistry 29, 10573–10576 (1990).
Herschlag, D. & Cech, T.R. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry 29, 10159–10171 (1990).
Zarrinkar, P.P., Wang, J. & Williamson, J.R. Slow folding kinetics of RNase P RNA. RNA 2, 564–573 (1996).
Pan, J., Thirumalai, D. & Woodson, S.A. Folding of RNA involves parallel pathways. J. Mol. Biol. 273, 7–13 (1997).
Esteban, J.A., Banerjee, A.R. & Burke, J.M. Kinetic mechanism of the hairpin ribozyme. Identification and characterization of two nonexchangeable conformations. J. Biol. Chem. 272, 13629–13639 (1997).
Pan, T. & Sosnick, T.R. Intermediates and kinetic traps in the folding of a large ribozyme revealed by circular dichroism and UV absorbance spectroscopies and catalytic activity. Nat. Struct. Biol. 4, 931–938 (1997).
Russell, R. & Herschlag, D. Probing the folding landscape of the Tetrahymena ribozyme: commitment to form the native conformation is late in the folding pathway. J. Mol. Biol. 308, 839–851 (2001).
Pichler, A. & Schroeder, R. Folding problems of the 5′ splice site containing the P1 stem of the group I thymidylate synthase intron: substrate binding inhibition in vitro and mis-splicing in vivo. J. Biol. Chem. 277, 17987–17993 (2002).
Chadalavada, D.M., Senchak, S.E. & Bevilacqua, P.C. The folding pathway of the genomic hepatitis delta virus ribozyme is dominated by slow folding of the pseudoknots. J. Mol. Biol. 317, 559–575 (2002).
Gerard, G.F., Fox, D.K., Nathan, M. & D'Alessio, J.M. Reverse transcriptase. The use of cloned Moloney murine leukemia virus reverse transcriptase to synthesize DNA from RNA. Mol. Biotechnol. 8, 61–77 (1997).
Das, R., Laederach, A., Pearlman, S.M., Herschlag, D. & Altman, R.B. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA 11, 344–354 (2005).
Stein, A. & Crothers, D.M. Conformational changes of transfer RNA. The role of magnesium(II). Biochemistry 15, 160–168 (1976).
Grilley, D., Soto, A.M. & Draper, D.E. Mg2+–RNA interaction free energies and their relationship to the folding of RNA tertiary structures. Proc. Natl. Acad. Sci. USA 103, 14003–14008 (2006).
Matsuura, M., Noah, J.W. & Lambowitz, A.M. Mechanism of maturase-promoted group II intron splicing. EMBO J. 20, 7259–7270 (2001).
Acknowledgements
We thank Yaqi Wan for assistance in preparing Figure 4 and Liz Doherty and Jennifer Doudna for early advice on footprinting and for sharing their protocol. Research in our labs is supported by grants from the NIH (R01-GM070456 to R.R. and R01-GM037951 to Alan M. Lambowitz) and the Welch Foundation (F-1563 to R.R.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tijerina, P., Mohr, S. & Russell, R. DMS footprinting of structured RNAs and RNA–protein complexes. Nat Protoc 2, 2608–2623 (2007). https://doi.org/10.1038/nprot.2007.380
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.380
This article is cited by
-
A chemical probe based on the PreQ1 metabolite enables transcriptome-wide mapping of binding sites
Nature Communications (2021)
-
The impact of various seed, accessibility and interaction constraints on sRNA target prediction- a systematic assessment
BMC Bioinformatics (2020)
-
The chemistry and applications of RNA 2′-OH acylation
Nature Reviews Chemistry (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.