Abstract
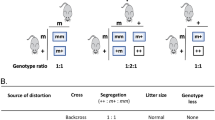
Transmission ratio distortion in the mouse is caused by several t-complex distorters (Tcds) acting in trans on the t-complex responder (Tcr)1,2,3,4. Tcds additively affect the flagellar movement of all spermatozoa derived from t/+ males; sperm carrying Tcr are rescued, resulting in an advantage for t sperm in fertilization. Here we show that Tagap1, a GTPase-activating protein, can act as a distorter. Tagap1 maps to the Tcd1 interval and has four t loci, which encode altered proteins including a C-terminally truncated form. Overexpression of wild-type Tagap1 in sperm cells phenocopied Tcd function, whereas a loss-of-function Tagap1 allele reduced the transmission rate of the t6 haplotype. The combined data strongly suggest that the t loci of Tagap1 produce Tcd1a. Our results unravel the molecular nature of a Tcd and demonstrate the importance of small G proteins in transmission ratio distortion in the mouse.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Silver, L.M. The peculiar journey of a selfish chromosome: mouse t haplotypes and meiotic drive. Trends Genet. 9, 250–254 (1993).
Lyon, M.F. Transmission ratio distortion in mice. Annu. Rev. Genet. 37, 393–408 (2003).
Lyon, M.F. Transmission ratio distortion in mouse t-haplotypes is due to multiple distorter genes acting on a responder locus. Cell 37, 621–628 (1984).
Silver, L.M. & Remis, D. Five of the nine genetically defined regions of mouse t haplotypes are involved in transmission ratio distortion. Genet. Res. 49, 51–56 (1987).
Katz, D.F., Erickson, R.P. & Nathanson, M. Beat frequency is bimodally distributed in spermatozoa from T/t12 mice. J. Exp. Zool. 210, 529–535 (1979).
Olds-Clarke, P. & Johnson, L.R. t haplotypes in the mouse compromise sperm flagellar function. Dev. Biol. 155, 14–25 (1993).
Herrmann, B.G., Koschorz, B., Wertz, K., McLaughlin, K.J. & Kispert, A. A protein kinase encoded by the t complex responder gene causes non-mendelian inheritance. Nature 402, 141–146 (1999).
Herrmann, B.G., Barlow, D.P. & Lehrach, H. A large inverted duplication allows homologous recombination between chromosomes heterozygous for the proximal t complex inversion. Cell 48, 813–825 (1987).
Kleene, K.C. Poly(A) shortening accompanies the activation of translation of five mRNAs during spermiogenesis in the mouse. Development 106, 367–373 (1989).
Gummere, G.R., McCormick, P.J. & Bennett, D. The influence of genetic background and the homologous chromosome 17 on t-haplotype transmission ratio distortion in mice. Genetics 114, 235–245 (1986).
Wennerberg, K. & Der, C.J. Rho-family GTPases: it's not only Rac and Rho (and I like it). J. Cell Sci. 117, 1301–1312 (2004).
Howard, T., Balogh, R., Overbeek, P. & Bernstein, K.E. Sperm-specific expression of angiotensin-converting enzyme (ACE) is mediated by a 91-base-pair promoter containing a CRE-like element. Mol. Cell. Biol. 13, 18–27 (1993).
Lyon, M.F. Deletion of mouse t-complex distorter-1 produces an effect like that of the t-form of the distorter. Genet. Res. 59, 27–33 (1992).
Lyon, M.F., Schimenti, J.C. & Evans, E.P. Narrowing the critical regions for mouse t complex transmission ratio distortion factors by use of deletions. Genetics 155, 793–801 (2000).
Merrill, C., Bayraktaroglu, L., Kusano, A. & Ganetzky, B. Truncated RanGAP encoded by the Segregation Distorter locus of Drosophila. Science 283, 1742–1745 (1999).
Kusano, A., Staber, C. & Ganetzky, B. Nuclear mislocalization of enzymatically active RanGAP causes segregation distortion in Drosophila. Dev. Cell 1, 351–361 (2001).
Kusano, A., Staber, C. & Ganetzky, B. Segregation distortion induced by wild-type RanGAP in Drosophila. Proc. Natl. Acad. Sci. USA 99, 6866–6870 (2002).
Hinsch, K.D. et al. ADP-ribosylation of Rho proteins inhibits sperm motility. FEBS Lett. 334, 32–36 (1993).
Nakamura, K. et al. Rhophilin, a small GTPase Rho-binding protein, is abundantly expressed in the mouse testis and localized in the principal piece of the sperm tail. FEBS Lett. 445, 9–13 (1999).
Fujita, A. et al. Ropporin, a sperm-specific binding protein of rhophilin, that is localized in the fibrous sheath of sperm flagella. J. Cell Sci. 113, 103–112 (2000).
Church, G.M. & Gilbert, W. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81, 1991–1995 (1984).
Dinkel, A. et al. Efficient generation of transgenic BALB/c mice using BALB/c embryonic stem cells. J. Immunol. Methods 223, 255–260 (1999).
Gilman, M. Ribonuclease Protection Assay (John Wiley & Sons, Inc., New York, 1997).
Ramirez-Solis, R., Davis, A.C. & Bradley, A. Gene targeting in embryonic stem cells. Methods Enzymol. 225, 855–878 (1993).
Chung, J.H., Whiteley, M. & Felsenfeld, G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74, 505–514 (1993).
Self, A.J. & Hall, A. Measurement of intrinsic nucleotide exchange and GTP hydrolysis rates. Methods Enzymol. 256, 67–76 (1995).
Acknowledgements
We thank M.F. Lyon for mice carrying the t haplotypes used in this study, D. Solter for long-term support, K. Wertz for the knockout allele of Tagaps in ES cells, B. Ledermann for BALB/c ES cells, D. Walther for Rho cDNA clones, M. Mallo for advice on ES-cell culture, B. Kanzler for pronuclear and ES-cell injections and L. Hartmann and the animal facilities of the Max Planck Institutes of Immunobiology and of Molecular Genetics for animal work. This project was supported by a grant from the Deutsche Forschungsgemeinschaft to B.G.H.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
Oligonucleotide primer sequences and PCR conditions used. (PDF 52 kb)
Rights and permissions
About this article
Cite this article
Bauer, H., Willert, J., Koschorz, B. et al. The t complex–encoded GTPase-activating protein Tagap1 acts as a transmission ratio distorter in mice. Nat Genet 37, 969–973 (2005). https://doi.org/10.1038/ng1617
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1617
This article is cited by
-
Non-Mendelian segregation and transmission drive of B chromosomes
Chromosome Research (2022)
-
Non-Mendelian transmission of accessory chromosomes in fungi
Chromosome Research (2022)
-
A meiotic driver alters sperm form and function in house mice: a possible example of spite
Chromosome Research (2022)
-
Mechanisms of meiotic drive in symmetric and asymmetric meiosis
Cellular and Molecular Life Sciences (2021)
-
TAGAP instructs Th17 differentiation by bridging Dectin activation to EPHB2 signaling in innate antifungal response
Nature Communications (2020)