Abstract
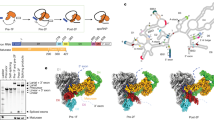
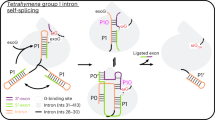
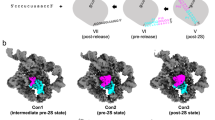
Although the importance of large noncoding RNAs is increasingly appreciated, our understanding of their structures and architectural dynamics remains limited. In particular, we know little about RNA folding intermediates and how they facilitate the productive assembly of RNA tertiary structures. Here, we report the crystal structure of an obligate intermediate that is required during the earliest stages of group II intron folding. Composed of domain 1 from the Oceanobacillus iheyensis group II intron (266 nucleotides), this intermediate retains native-like features but adopts a compact conformation in which the active site cleft is closed. Transition between this closed and the open (native) conformation is achieved through discrete rotations of hinge motifs in two regions of the molecule. The open state is then stabilized by sequential docking of downstream intron domains, suggesting a 'first come, first folded' strategy that may represent a generalizable pathway for assembly of large RNA and ribonucleoprotein structures.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Treiber, D.K. & Williamson, J.R. Beyond kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 11, 309–314 (2001).
Woodson, S.A. Recent insights on RNA folding mechanisms from catalytic RNA. Cell. Mol. Life Sci. 57, 796–808 (2000).
Sosnick, T.R. & Pan, T. RNA folding: models and perspectives. Curr. Opin. Struct. Biol. 13, 309–316 (2003).
Su, L.J., Waldsich, C. & Pyle, A.M. An obligate intermediate along the slow folding pathway of a group II intron ribozyme. Nucleic Acids Res. 33, 6674–6687 (2005).
Woodson, S.A. Compact intermediates in RNA folding. Annu. Rev. Biophys. 39, 61–77 (2010).
Bryngelson, J.D., Onuchic, J.N., Socci, N.D. & Wolynes, P.G. Funnels, pathways, and the energy landscape of protein folding: a synthesis. Proteins 21, 167–195 (1995).
Kim, P.S. & Baldwin, R.L. Intermediates in the folding reactions of small proteins. Annu. Rev. Biochem. 59, 631–660 (1990).
Dyson, H.J. & Wright, P.E. Peptide conformation and protein-folding. Curr. Opin. Struct. Biol. 3, 60–65 (1993).
Pyle, A.M. The tertiary structure of group II introns: implications for biological function and evolution. Crit. Rev. Biochem. Mol. Biol. 45, 215–232 (2010).
Pyle, A.M. & Lambowitz, A.M. Group II introns: ribozymes that splice RNA and invade DNA. in The RNA World 3rd edn. (ed. Gesteland, R.F.) 469–505 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, USA, 2006).
Pyle, A.M., Fedorova, O. & Waldsich, C. Folding of group II introns: a model system for large, multidomain RNAs? Trends Biochem. Sci. 32, 138–145 (2007).
Krasilnikov, A.S., Xiao, Y., Pan, T. & Mondragon, A. Basis for structural diversity in homologous RNAs. Science 306, 104–107 (2004).
Cate, J.H. et al. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science 273, 1678–1685 (1996).
Marcia, M. & Pyle, A.M. Visualizing group II intron catalysis through the stages of splicing. Cell 151, 497–507 (2012).
Toor, N., Keating, K.S., Taylor, S.D. & Pyle, A.M. Crystal structure of a self-spliced group II intron. Science 320, 77–82 (2008).
Coonrod, L.A., Lohman, J.R. & Berglund, J.A. Utilizing the GAAA tetraloop/receptor to facilitate crystal packing and determination of the structure of a CUG RNA helix. Biochemistry 51, 8330–8337 (2012).
Marcia, M. et al. Solving nucleic acid structures by molecular replacement: examples from group II intron studies. Acta Crystallogr. D Biol. Crystallogr. 69, 2174–2185 (2013).
Robertson, M.P. & Scott, W.G. A general method for phasing novel complex RNA crystal structures without heavy-atom derivatives. Acta Crystallogr. D Biol. Crystallogr. D64, 738–744 (2008).
Marcia, M. & Pyle, A.M. Principles of ion recognition in RNA: insights from the group II intron structures. RNA 20, 516–527 (2014).
Qin, P.Z. & Pyle, A.M. Stopped-flow fluorescence spectroscopy of a group II intron ribozyme reveals that domain 1 is an independent folding unit with a requirement for specific Mg2+ ions in the tertiary structure. Biochemistry 36, 4718–4730 (1997).
Michels, W.J. Jr. & Pyle, A.M. Conversion of a group II intron into a new multiple-turnover ribozyme that selectively cleaves oligonucleotides: elucidation of reaction mechanism and structure/function relationships. Biochemistry 34, 2965–2977 (1995).
Drenth, J. Theory of X-ray diffraction by a crystal. Principles of Protein X-Ray Crystallography Chapter 4, 64–108 (Springer, 2007).
Mohan, S., Donohue, J.P. & Noller, H.F. Molecular mechanics of 30S subunit head rotation. Proc. Natl. Acad. Sci. USA 111, 13325–13330 (2014).
Toor, N. et al. Tertiary architecture of the Oceanobacillus iheyensis group II intron. RNA 16, 57–69 (2010).
Keating, K.S., Humphris, E.L. & Pyle, A.M. A new way to see RNA. Q. Rev. Biophys. 44, 433–466 (2011).
Duarte, C.M., Wadley, L.M. & Pyle, A.M. RNA structure comparison, motif search and discovery using a reduced representation of RNA conformational space. Nucleic Acids Res. 31, 4755–4761 (2003).
Duarte, C.M. & Pyle, A.M. Stepping through an RNA structure: a novel approach to conformational analysis. J. Mol. Biol. 284, 1465–1478 (1998).
Fedorova, O. & Pyle, A.M. The brace for a growing scaffold: Mss116 protein promotes RNA folding by stabilizing an early assembly intermediate. J. Mol. Biol. 422, 347–365 (2012).
Guo, F., Gooding, A.R. & Cech, T.R. Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site. Mol. Cell 16, 351–362 (2004).
Juneau, K., Podell, E., Harrington, D.J. & Cech, T.R. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA–solvent interactions. Structure 9, 221–231 (2001).
Waldsich, C. & Pyle, A.M. A kinetic intermediate that regulates proper folding of a group II intron RNA. J. Mol. Biol. 375, 572–580 (2008).
Konforti, B.B., Liu, Q. & Pyle, A.M. A map of the binding site for catalytic domain 5 in the core of a group II intron ribozyme. EMBO J. 17, 7105–7117 (1998).
Torres-Larios, A., Swinger, K.K., Krasilnikov, A.S., Pan, T. & Mondragon, A. Crystal structure of the RNA component of bacterial ribonuclease P. Nature 437, 584–587 (2005).
Baird, N.J., Westhof, E., Qin, H., Pan, T. & Sosnick, T.R. Structure of a folding intermediate reveals the interplay between core and peripheral elements in RNA folding. J. Mol. Biol. 352, 712–722 (2005).
Pan, T. Folding of bacterial RNase P RNA. in Protein Reviews Vol. Ribonuclease P (eds. Liu, F. & Altman, S.) 79–91 (Springer, 2010).
Bailor, M.H., Mustoe, A.M., Brooks, C.L. III. & Al-Hashimi, H.M. Topological constraints: using RNA secondary structure to model 3D conformation, folding pathways, and dynamic adaptation. Curr. Opin. Struct. Biol. 21, 296–305 (2011).
Bailor, M.H., Sun, X. & Al-Hashimi, H.M. Topology links RNA secondary structure with global conformation, dynamics, and adaptation. Science 327, 202–206 (2010).
Chu, V.B. et al. Do conformational biases of simple helical junctions influence RNA folding stability and specificity? RNA 15, 2195–2205 (2009).
Behrouzi, R., Roh, J.H., Kilburn, D., Briber, R.M. & Woodson, S.A. Cooperative tertiary interaction network guides RNA folding. Cell 149, 348–357 (2012).
Johnson, T.H., Tijerina, P., Chadee, A.B., Herschlag, D. & Russell, R. Structural specificity conferred by a group I RNA peripheral element. Proc. Natl. Acad. Sci. USA 102, 10176–10181 (2005).
Pan, T. & Sosnick, T. RNA folding during transcription. Annu. Rev. Biophys. Biomol. Struct. 35, 161–175 (2006).
Somarowthu, S. et al. HOTAIR forms an intricate and modular secondary structure. Mol. Cell 58, 353–361 (2015).
Chillon, I. et al. Native purification and analysis of long RNAs. Methods Enzymol. 558, 3–37 (2015).
Hughson, F.M., Wright, P.E. & Baldwin, R.L. Structural characterization of a partly folded apomyoglobin intermediate. Science 249, 1544–1548 (1990).
Bhutani, N. & Udgaonkar, J.B. Folding subdomains of thioredoxin characterized by native-state hydrogen exchange. Protein Sci. 12, 1719–1731 (2003).
Llinás, M. & Marqusee, S. Subdomain interactions as a determinant in the folding and stability of T4 lysozyme. Protein Sci. 7, 96–104 (1998).
Wu, L.C., Grandori, R. & Carey, J. Autonomous subdomains in protein folding. Protein Sci. 3, 369–371 (1994).
Uzawa, T. et al. Hierarchical folding mechanism of apomyoglobin revealed by ultra-fast H/D exchange coupled with 2D NMR. Proc. Natl. Acad. Sci. USA 105, 13859–13864 (2008).
Chworos, A. et al. Building programmable jigsaw puzzles with RNA. Science 306, 2068–2072 (2004).
Han, D. et al. DNA origami with complex curvatures in three-dimensional space. Science 332, 342–346 (2011).
Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Winn, M.D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
Sheldrick, G.M. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D Biol. Crystallogr. 66, 479–485 (2010).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Keating, K.S. & Pyle, A.M. RCrane: semi-automated RNA model building. Acta Crystallogr. D Biol. Crystallogr. 68, 985–995 (2012).
Karplus, P.A. & Diederichs, K. Assessing and maximizing data quality in macromolecular crystallography. Curr. Opin. Struct. Biol. 34, 60–68 (2015).
Krebs, W.G. & Gerstein, M. The morph server: a standardized system for analyzing and visualizing macromolecular motions in a database framework. Nucleic Acids Res. 28, 1665–1675 (2000).
Lu, X.J. & Olson, W.K. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 31, 5108–5121 (2003).
Schneider, B., Gelly, J.C., de Brevern, A.G. & Cerny, J. Local dynamics of proteins and DNA evaluated from crystallographic B factors. Acta Crystallogr. D Biol. Crystallogr. 70, 2413–2419 (2014).
Acknowledgements
We would like to thank S. Somarowthu for constructive discussions and O. Fedorova, S. Somarowthu and T. Dickey for reading the manuscript. C.Z. is supported by Gruber Science Fellowship. A.M.P. is a Howard Hughes Medical Institute Investigator. This work is supported by the US National Institute of Health (NIH) (RO1GM50313) and is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the NIH (P41 GM103403). The Pilatus 6M detector on 24-ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357.
Author information
Authors and Affiliations
Contributions
C.Z. designed the constructs and conducted all the experiments; C.Z., K.R.R. and M.M. collected X-ray diffraction data; and C.Z., K.R.R. and M.M. solved the structure. C.Z. analyzed the structure. A.M.P. and M.M. directed the research. A.M.P. and C.Z. wrote the manuscript, and M.M. and K.R.R. contributed to writing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Table 1 and Supplementary Figures 1–8. (PDF 0 kb)
Morphing between D1iso and D1full and subsequent docking of D2–D5.
This movie starts with a 360° rotation of the D1iso. Subsequently, D1iso opens up to D1full, then D1full closes to D1iso. This process is repeated one more time with hinge residues located at 5′H and 3′H colored in green and pink. Finally, D1iso opens to D1full for one more time, then downstream domains D2–D5 dock sequentially. D1–D5 are colored in grey, blue, wheat, yellow and red respectively. The procedure used to produce this video is described in Online Methods section of the article. (MOV 24403 kb)
Rights and permissions
About this article
Cite this article
Zhao, C., Rajashankar, K., Marcia, M. et al. Crystal structure of group II intron domain 1 reveals a template for RNA assembly. Nat Chem Biol 11, 967–972 (2015). https://doi.org/10.1038/nchembio.1949
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1949
This article is cited by
-
Visualizing group II intron dynamics between the first and second steps of splicing
Nature Communications (2020)
-
Visualizing the functional 3D shape and topography of long noncoding RNAs by single-particle atomic force microscopy and in-solution hydrodynamic techniques
Nature Protocols (2020)
-
A clear path to RNA catalysis
Nature Chemical Biology (2015)