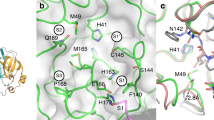
Abstract
We identified small-molecule dimer disruptors that inhibit an essential dimeric protease of human Kaposi's sarcoma–associated herpesvirus (KSHV) by screening an α-helical mimetic library. Next, we synthesized a second generation of low-micromolar inhibitors with improved potency and solubility. Complementary methods including size exclusion chromatography and 1H-13C HSQC titration using selectively labeled 13C-Met samples revealed that monomeric protease is enriched in the presence of inhibitor. 1H-15N HSQC titration studies mapped the inhibitor binding site to the dimer interface, and mutagenesis studies targeting this region were consistent with a mechanism where inhibitor binding prevents dimerization through the conformational selection of a dynamic intermediate. These results validate the interface of herpesvirus proteases and other similar oligomeric interactions as suitable targets for the development of small-molecule inhibitors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Jones, S. & Thornton, J.M. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. USA 93, 13–20 (1996).
Hopkins, A.L. & Groom, C.R. The druggable genome. Nat. Rev. Drug Discov. 1, 727–730 (2002).
Lo Conte, L., Chothia, C. & Janin, J. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 285, 2177–2198 (1999).
Berg, T. Modulation of protein-protein interactions with small organic molecules. Angew. Chem. Int. Edn. Engl. 42, 2462–2481 (2003).
Wells, J.A. & McClendon, C.L. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature 450, 1001–1009 (2007).
Tse, C. et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 68, 3421–3428 (2008).
Fields, B.N. et al. Fields Virology 3177 (Lippincott Williams & Wilkins, Philadelphia, 2006).
Gopalsamy, A. et al. Design and syntheses of 1,6-naphthalene derivatives as selective HCMV protease inhibitors. J. Med. Chem. 47, 1893–1899 (2004).
Gao, M. et al. The protease of herpes simplex virus type 1 is essential for functional capsid formation and viral growth. J. Virol. 68, 3702–3712 (1994).
Preston, V.G., Coates, J.A. & Rixon, F.J. Identification and characterization of a herpes simplex virus gene product required for encapsidation of virus DNA. J. Virol. 45, 1056–1064 (1983).
Sheaffer, A.K. et al. Evidence for controlled incorporation of herpes simplex virus type 1 UL26 protease into capsids. J. Virol. 74, 6838–6848 (2000).
Weinheimer, S.P. et al. Autoproteolysis of herpes simplex virus type 1 protease releases an active catalytic domain found in intermediate capsid particles. J. Virol. 67, 5813–5822 (1993).
Welch, A.R., Woods, A.S., McNally, L.M., Cotter, R.J. & Gibson, W. A herpesvirus maturational proteinase, assemblin: identification of its gene, putative active site domain, and cleavage site. Proc. Natl. Acad. Sci. USA 88, 10792–10796 (1991).
Borthwick, A.D. et al. Design and synthesis of pyrrolidine-5,5-trans-lactams (5-oxohexahydropyrrolo[3,2-b]pyrroles) as novel mechanism-based inhibitors of human cytomegalovirus protease. 2. Potency and chirality. J. Med. Chem. 45, 1–18 (2002).
Borthwick, A.D. et al. Pyrrolidine-5,5-trans-lactams as novel mechanism-based inhibitors of human cytomegalovirus protease. Part 3: potency and plasma stability. Bioorg. Med. Chem. Lett. 12, 1719–1722 (2002).
Borthwick, A.D. et al. Design and synthesis of monocyclic beta-lactams as mechanism-based inhibitors of human cytomegalovirus protease. Bioorg. Med. Chem. Lett. 8, 365–370 (1998).
Ogilvie, W. et al. Peptidomimetic inhibitors of the human cytomegalovirus protease. J. Med. Chem. 40, 4113–4135 (1997).
Waxman, L. & Darke, P.L. The herpesvirus proteases as targets for antiviral chemotherapy. Antivir. Chem. Chemother. 11, 1–22 (2000).
Batra, R., Khayat, R. & Tong, L. Molecular mechanism for dimerization to regulate the catalytic activity of human cytomegalovirus protease. Nat. Struct. Biol. 8, 810–817 (2001).
Buisson, M. et al. Functional determinants of the Epstein-Barr virus protease. J. Mol. Biol. 311, 217–228 (2001).
Darke, P.L. et al. Active human cytomegalovirus protease is a dimer. J. Biol. Chem. 271, 7445–7449 (1996).
Margosiak, S.A., Vanderpool, D.L., Sisson, W., Pinko, C. & Kan, C.C. Dimerization of the human cytomegalovirus protease: kinetic and biochemical characterization of the catalytic homodimer. Biochemistry 35, 5300–5307 (1996).
Nomura, A.M., Marnett, A.B., Shimba, N., Dotsch, V. & Craik, C.S. Induced structure of a helical switch as a mechanism to regulate enzymatic activity. Nat. Struct. Mol. Biol. 12, 1019–1020 (2005).
Pray, T.R., Reiling, K.K., Demirjian, B.G. & Craik, C.S. Conformational change coupling the dimerization and activation of KSHV protease. Biochemistry 41, 1474–1482 (2002).
Reiling, K.K., Pray, T.R., Craik, C.S. & Stroud, R.M. Functional consequences of the Kaposi's sarcoma-associated herpesvirus protease structure: regulation of activity and dimerization by conserved structural elements. Biochemistry 39, 12796–12803 (2000).
Schmidt, U. & Darke, P.L. Dimerization and activation of the herpes simplex virus type 1 protease. J. Biol. Chem. 272, 7732–7735 (1997).
Nomura, A.M., Marnett, A.B., Shimba, N., Dotsch, V. & Craik, C.S. One functional switch mediates reversible and irreversible inactivation of a herpesvirus protease. Biochemistry 45, 3572–3579 (2006).
Marnett, A.B., Nomura, A.M., Shimba, N., Ortiz de Montellano, P.R. & Craik, C.S. Communication between the active sites and dimer interface of a herpesvirus protease revealed by a transition-state inhibitor. Proc. Natl. Acad. Sci. USA 101, 6870–6875 (2004).
Buisson, M. et al. The crystal structure of the Epstein-Barr virus protease shows rearrangement of the processed C terminus. J. Mol. Biol. 324, 89–103 (2002).
Qiu, X. et al. Unique fold and active site in cytomegalovirus protease. Nature 383, 275–279 (1996).
Qiu, X. et al. Crystal structure of varicella-zoster virus protease. Proc. Natl. Acad. Sci. USA 94, 2874–2879 (1997).
Shieh, H.S. et al. Three-dimensional structure of human cytomegalovirus protease. Nature 383, 279–282 (1996).
Tong, L. et al. A new serine-protease fold revealed by the crystal structure of human cytomegalovirus protease. Nature 383, 272–275 (1996).
Hoog, S.S. et al. Active site cavity of herpesvirus proteases revealed by the crystal structure of herpes simplex virus protease/inhibitor complex. Biochemistry 36, 14023–14029 (1997).
Pray, T.R., Nomura, A.M., Pennington, M.W. & Craik, C.S. Auto-inactivation by cleavage within the dimer interface of Kaposi's sarcoma-associated herpesvirus protease. J. Mol. Biol. 289, 197–203 (1999).
Shimba, N., Nomura, A.M., Marnett, A.B. & Craik, C.S. Herpesvirus protease inhibition by dimer disruption. J. Virol. 78, 6657–6665 (2004).
Lu, F. et al. Proteomimetic libraries: design, synthesis, and evaluation of p53–MDM2 interaction inhibitors. J. Comb. Chem. 8, 315–325 (2006).
Backes, B.J., Harris, J.L., Leonetti, F., Craik, C.S. & Ellman, J.A. Synthesis of positional-scanning libraries of fluorogenic peptide substrates to define the extended substrate specificity of plasmin and thrombin. Nat. Biotechnol. 18, 187–193 (2000).
Lazic, A., Goetz, D.H., Nomura, A.M., Marnett, A.B. & Craik, C.S. Substrate modulation of enzyme activity in the herpesvirus protease family. J. Mol. Biol. 373, 913–923 (2007).
Feng, B.Y. & Shoichet, B.K. A detergent-based assay for the detection of promiscuous inhibitors. Nat. Protoc. 1, 550–553 (2006).
Lee, G.M. & Craik, C.S. Trapping moving targets with small molecules. Science 324, 213–215 (2009).
Kussie, P.H. et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274, 948–953 (1996).
Chen, P. et al. Structure of the human cytomegalovirus protease catalytic domain reveals a novel serine protease fold and catalytic triad. Cell 86, 835–843 (1996).
Wu, T.D. et al. Mutation patterns and structural correlates in human immunodeficiency virus type 1 protease following different protease inhibitor treatments. J. Virol. 77, 4836–4847 (2003).
Hardy, J.A., Lam, J., Nguyen, J.T., O'Brien, T. & Wells, J.A. Discovery of an allosteric site in the caspases. Proc. Natl. Acad. Sci. USA 101, 12461–12466 (2004).
Scheer, J.M., Romanowski, M.J. & Wells, J.A. A common allosteric site and mechanism in caspases. Proc. Natl. Acad. Sci. USA 103, 7595–7600 (2006).
Bannwarth, L. & Reboud-Ravaux, M. An alternative strategy for inhibiting multidrug-resistant mutants of the dimeric HIV-1 protease by targeting the subunit interface. Biochem. Soc. Trans. 35, 551–554 (2007).
Lee, S.G. & Chmielewski, J. Rapid synthesis and in situ screening of potent HIV-1 protease dimerization inhibitors. Chem. Biol. 13, 421–426 (2006).
Unal, A. et al. The protease and the assembly protein of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8). J. Virol. 71, 7030–7038 (1997).
Brignole, E.J. & Gibson, W. Enzymatic activities of human cytomegalovirus maturational protease assemblin and its precursor (pPR, pUL80a) are comparable: [corrected] maximal activity of pPR requires self-interaction through its scaffolding domain. J. Virol. 81, 4091–4103 (2007).
Acknowledgements
The authors thank W. Gibson (The Johns Hopkins University School of Medicine) for providing us with the CMV protease expression plasmid and for valuable feedback on the manuscript. This work was funded by US National Institutes of Health grants T32 GMO7810 and AIO67423 (C.S.C.) and by the American Lebanese and Syrian Associated Charities and the St. Jude Children's Research Hospital (R.K.G.). We also thank the University of California, San Francisco–Gladstone Institute for Virology and Immunology/Center for AIDS Research for the Clinical Science Pilot Award (P30-AI027763 to G.M.L.).
Author information
Authors and Affiliations
Contributions
T.S., G.M.L., A.L., C.S.C. and R.K.G. designed research; T.S., G.M.L. and A.L. carried out research; L.A.A., P.V. and C.M.R. performed chemical synthesis; T.S., G.M.L., A.L., C.S.C. and R.K.G. analyzed and interpreted data; and T.S., C.S.C. and R.K.G. prepared the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Scheme 1, Supplementary Tables 1 and 2, and Supplementary Methods (PDF 481 kb)
Rights and permissions
About this article
Cite this article
Shahian, T., Lee, G., Lazic, A. et al. Inhibition of a viral enzyme by a small-molecule dimer disruptor. Nat Chem Biol 5, 640–646 (2009). https://doi.org/10.1038/nchembio.192
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.192
This article is cited by
-
Allosteric Site Inhibitor Disrupting Auto-Processing of Malarial Cysteine Proteases
Scientific Reports (2018)
-
Inhibition of α-helix-mediated protein–protein interactions using designed molecules
Nature Chemistry (2013)
-
Defining an allosteric circuit in the cysteine protease domain of Clostridium difficile toxins
Nature Structural & Molecular Biology (2011)
-
Helix-mediated protein–protein interactions as targets for intervention using foldamers
Amino Acids (2011)
-
Emerging principles in protease-based drug discovery
Nature Reviews Drug Discovery (2010)