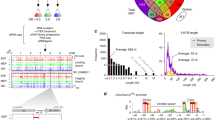
Abstract
Estimates of the total number of bacterial species1,2,3 indicate that existing DNA sequence databases carry only a tiny fraction of the total amount of DNA sequence space represented by this division of life. Indeed, environmental DNA samples have been shown to encode many previously unknown classes of proteins4 and RNAs5. Bioinformatics searches6,7,8,9,10 of genomic DNA from bacteria commonly identify new noncoding RNAs (ncRNAs)10,11,12 such as riboswitches13,14. In rare instances, RNAs that exhibit more extensive sequence and structural conservation across a wide range of bacteria are encountered15,16. Given that large structured RNAs are known to carry out complex biochemical functions such as protein synthesis and RNA processing reactions, identifying more RNAs of great size and intricate structure is likely to reveal additional biochemical functions that can be achieved by RNA. We applied an updated computational pipeline17 to discover ncRNAs that rival the known large ribozymes in size and structural complexity or that are among the most abundant RNAs in bacteria that encode them. These RNAs would have been difficult or impossible to detect without examining environmental DNA sequences, indicating that numerous RNAs with extraordinary size, structural complexity, or other exceptional characteristics remain to be discovered in unexplored sequence space.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Whitman, W. B., Coleman, D. C. & Wiebe, W. J. Prokaryotes: the unseen majority. Proc. Natl Acad. Sci. USA 95, 6578–6583 (1998)
Curtis, T. P., Sloan, W. T. & Scannell, J. W. Estimating prokaryotic diversity and its limits. Proc. Natl Acad. Sci. USA 99, 10494–10499 (2002)
Bent, S. J. & Forney, L. J. The tragedy of the uncommon: understanding limitations in the analysis of microbial diversity. ISME J. 2, 689–695 (2008)
Yooseph, S. et al. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 5, e16 (2007)
Shi, Y., Tyson, G. W. & DeLong, E. F. Metatranscriptomics reveals unique microbial small RNAs in the ocean's water column. Nature 459, 266–269 (2009)
Gelfand, M. S., Mironov, A. A., Jomantas, J., Kozlov, Y. I. & Perumov, D. A. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet. 15, 439–442 (1999)
Rivas, E. & Eddy, S. R. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics 2, 8 (2001)
Wassarman, K. M., Repoila, F., Rosenow, C., Storz, G. & Gottesman, S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 15, 1637–1651 (2001)
Barrick, J. E. et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl Acad. Sci. USA 101, 6421–6426 (2004)
Yao, Z. et al. A computational pipeline for high-throughput discovery of cis-regulatory noncoding RNA in prokaryotes. PLOS Comput. Biol. 3, e126 (2007)
Weinberg, Z. et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 35, 4809–4819 (2007)
Meyer, M. M. et al. Identification of candidate structured RNAs in the marine organism ‘Candidatus Pelagibacter ubique’. BMC Genomics 10, 268 (2009)
Montange, R. K. & Batey, R. T. Riboswitches: emerging themes in RNA structure and function. Annu. Rev. Biophys. 37, 117–133 (2008)
Roth, A. & Breaker, R. R. The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 78, 305–334 (2009)
Barrick, J. E., Sudarsan, N., Weinberg, Z., Ruzzo, W. L. & Breaker, R. R. 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA 11, 774–784 (2005)
Puerta-Fernandez, E., Barrick, J. E., Roth, A. & Breaker, R. R. Identification of a large noncoding RNA in extremophilic eubacteria. Proc. Natl Acad. Sci. USA 103, 19490–19495 (2006)
Tseng, H. H., Weinberg, Z., Gore, J., Breaker, R. R. & Ruzzo, W. L. Finding non-coding RNAs through genome-scale clustering. J. Bioinform. Comput. Biol. 7, 373–388 (2009)
Michel, F. & Westhof, E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 216, 585–610 (1990)
Pace, N. R., Thomas, B. C. & Woese, C. R. in The RNA World 2nd edn (eds Gesteland, R. F., Cech, T. R. & Atkins, J. F.) Ch. 4 113–141 (Cold Spring Harbor Laboratory Press, 1999)
Toor, N., Keating, K. S. & Pyle, A. M. Structural insights into RNA splicing. Curr. Opin. Struct. Biol. 19, 260–266 (2009)
Rusch, D. B. et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5, e77 (2007)
Raya, R. R. & Hébert, E. M. in Bacteriophages: Methods and Protocols Vol. 1 (ed. Clokie, M. R. J.) (Humana, 2009)
Stoddard, B. L. Homing endonuclease structure and function. Q. Rev. Biophys. 38, 49–95 (2005)
Lambowitz, A. M. & Zimmerly, S. Mobile group II introns. Annu. Rev. Genet. 38, 1–35 (2004)
Wassarman, K. M., Zhang, A. & Storz, G. Small RNAs in Escherichia coli . Trends Microbiol. 7, 37–45 (1999)
Frias-Lopez, J. et al. Microbial community gene expression in ocean surface waters. Proc. Natl Acad. Sci. USA 105, 3805–3810 (2008)
Wassarman, K. M. 6S RNA: a regulator of transcription. Mol. Microbiol. 65, 1425–1431 (2007)
Pichon, C. & Felden, B. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl Acad. Sci. USA 102, 14249–14254 (2005)
Altuvia, S., Weinstein-Fischer, D., Zhang, A., Postow, L. & Storz, G. A. Small, Stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell 90, 43–53 (1997)
Yao, Z., Weinberg, Z. & Ruzzo, W. L. CMfinder—a covariance model based RNA motif finding algorithm. Bioinformatics 22, 445–452 (2006)
Weinberg, Z. & Ruzzo, W. L. Sequence-based heuristics for faster annotation of non-coding RNA families. Bioinformatics 22, 35–39 (2006)
Eddy, S. R. & Durbin, R. RNA Sequence Analysis Using Covariance Models. Nucleic Acids Res. 22, 2079–2088 (1994)
Klein, R. J. & Eddy, S. R. RSEARCH: finding homologs of single structured RNA sequences. BMC Bioinformatics 4, 44 (2003)
Knudsen, B. & Hein, J. Pfold: RNA secondary structure prediction using stochastic context-free grammars. Nucleic Acids Res. 31, 3423–3428 (2003)
Yao, Z. Genome scale search of noncoding RNAs: bacteria to vertebrates. Dissertation, Univ. of Washington (2008)
Pruitt, K., Tatusova, T. & Maglott, D. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33, D501–D504 (2005)
Tyson, G. W. et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428, 37–43 (2004)
Tringe, S. G. et al. Comparative metagenomics of microbial communities. Science 308, 554–557 (2005)
Gill, S. R. et al. Metagenomic analysis of the human distal gut microbiome. Science 312, 1355–1359 (2006)
Kurokawa, K. et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 14, 169–181 (2007)
Turnbaugh, P. J. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006)
Woyke, T. et al. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature 443, 950–955 (2006)
Martín, H. G. et al. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nature Biotechnol. 24, 1263–1269 (2006)
Warnecke, F. et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450, 560–565 (2007)
Konstantidinis, K. T. et al. Comparative metagenomic analysis of a microbial community residing at a depth of 4,000 meters at station ALOHA in the North Pacific subtropical gyre. Appl. Environ. Microbiol. 75, 5345–5355 (2009)
Venter, J. C. et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74 (2004)
Noguchi, H., Park, J. & Takagi, T. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 34, 5623–5630 (2006)
Markowitz, V. M. et al. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res. 36, D534–D538 (2008)
Marchler-Bauer, A. et al. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 33, D192–D196 (2005)
Gardner, P. P. et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 37, D136–D140 (2009)
Liu, C. et al. NONCODE: an integrated knowledge database of non-coding RNAs. Nucleic Acids Res. 33, D112–D115 (2005)
Gutell, R. R., Larsen, N. & Woese, C. R. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol. Rev. 58, 10–26 (1994)
Dai, L. & Zimmerly, S. Compilation and analysis of group II intron insertions in bacterial genomes: evidence for retroelement behavior. Nucleic Acids Res. 30, 1091–1102 (2002)
Boudvillain, M. & Pyle, A. M. Defining functional groups, core structural features and inter-domain tertiary contacts essential for group II intron self-splicing: a NAIM analysis. EMBO J. 17, 7091–7104 (1998)
Toor, N., Hausner, G. & Zimmerly, S. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA 7, 1142–1152 (2001)
Haas, E. S., Brown, J. W., Pitulle, C. & Pace, N. R. Further perspective on the catalytic core and secondary structure of ribonuclease P RNA. Proc. Natl Acad. Sci. USA 91, 2527–2531 (1994)
Zwieb, C., Wower, I. & Wower, J. Comparative sequence analysis of tmRNA. Nucleic Acids Res. 27, 2063–2071 (1999)
Barrick, J. E. & Breaker, R. R. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 8, R239 (2007)
Guindon, S. & Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 (2003)
Cazenave, C. & Uhlenbeck, O. C. RNA-template-directed RNA synthesis by T7 polymerase. Proc. Natl Acad. Sci. USA 91, 6972–6976 (1994)
Wu, T., Ogilvie, T. T. & Pon, R. T. Prevention of chain cleavage in the chemical synthesis of 2′ silylated oligoribonucleotides. Nucleic Acids Res. 17, 3501–3517 (1989)
Regulski, E. E. & Breaker, R. R. in Methods in Molecular Biology Vol. 419 Post-Transcriptional Gene Regulation (ed. Wilusz, J.) (Humana, 2008)
Acknowledgements
We thank N. Carriero and R. Bjornson for assisting our use of the Yale Life Sciences High Performance Computing Center (NIH grant RR19895-02), T. Gruczka for advice and assistance in ocean water collection, J. Yang for assistance with the analysis of the dct-1 motif, D. Rodrigues for E. sibiricum, D. Bryant for A. maxima genomic DNA and P. O’Donoghue, M. Hammond, N. Sudarsan, S. Li, J. Barrick, Z. Yao, W. L. Ruzzo and E. Tseng for advice. J.P. and M.M.M. were supported by postdoctoral fellowships from the Canadian Institutes of Health Research and National Institutes of Health, respectively. R.R.B. is a Howard Hughes Medical Institute Investigator.
Author Contributions Z.W. and R.R.B. conceived the study and R.R.B. supervised the research. Z.W. created bioinformatics scripts and prepared RNA sequence alignments. J.P. conducted GOLLD and IMES RNA experiments. M.M.M. conducted GOLLD RACE and HEARO RNA experiments. Z.W. and R.R.B. wrote the manuscript, and all authors participated in editing.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Notes, Supplementary Tables 1-3, Supplementary Figures 1-11 with Legends and Supplementary References. (PDF 1207 kb)
Supplementary Data 1
This file presents detailed data on GOLLD, HEARO and IMES RNAs in printable format. For each RNA class, the organisms containing representatives are listed, and the nucleotide coordinates and genes surrounding each representative are depicted. The file also contains a full multiple-sequence alignment with consensus secondary structure for each RNA class. Also included are proposed alignments of regions of the 5' half of GOLLD RNA in Streptococus species, and a smaller structure that is more broadly detected. (PDF 3544 kb)
Supplementary Data 2
This compressed archive file houses multiple-sequence alignments in machine-readable format. The alignments were presented in printable form in Supplementary Data 1. The alignments include additional annotation used to generate drawings and printable data, as well as sequences that flank the RNAs. Each alignment is stored in "Stockholm" text format (http://en.wikipedia.org/wiki/Stockholm_format). The Stockholm files can be extracted from the .tar.gz format archive using programs such as WinZip (Windows), StuffIt Expander (Mac) or the tar/gzip commands (UNIX). (TAR 2050 kb)
Supplementary Data 3
This compressed archive file houses multiple-sequence alignments in Stockholm text format (http://en.wikipedia.org/wiki/Stockholm_format). However, annotation beyond the consensus secondary structure and flanking sequence is not included. The Stockholm files can be extracted from the .tar.gz format archive using programs such as WinZip (Windows), StuffIt Expander (Mac) or the tar/gzip commands (UNIX). (TAR 580 kb)
Rights and permissions
About this article
Cite this article
Weinberg, Z., Perreault, J., Meyer, M. et al. Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis . Nature 462, 656–659 (2009). https://doi.org/10.1038/nature08586
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature08586
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.