Abstract
Prader–Willi syndrome (PWS) is primarily caused by deletions involving the paternally derived imprinted region at chromosome 15q11.2–q13 and maternal uniparental disomy 15 (upd(15)mat). The underlying mechanisms for upd(15)mat include trisomy rescue (TR), gamete complementation (GC), monosomy rescue and post-fertilization mitotic error, and TR/GC is mediated by non-disjunction at maternal meiosis 1 (M1) or meiosis 2 (M2). Of these factors involved in the development of upd(15)mat, M1 non-disjunction is a maternal age-dependent phenomenon. We studied 117 Japanese patients with PWS and identified deletions in 84 patients (Deletion group) and TR/GC type upd(15)mat through M1 non-disjunction in 15 patients (TR/GC (M1) group), together with other types of abnormalities. Maternal age was significantly higher in TR/GC (M1) group than in Deletion group (median (range), 37 (35–45) versus 30 (19–42); P=1.0 × 10−7). Furthermore, delayed childbearing age became obvious since the year 2003 in Japan, and relative frequency of TR/GC (M1) group was significantly larger in patients born since the year 2003 than in those born until the year 2002. The results imply that the advanced maternal age at childbirth is a predisposing factor for the development of upd(15)mat because of increased M1 errors.
Similar content being viewed by others
Introduction
Prader–Willi syndrome (PWS) is a developmental disorder associated with various dysmorphic, neurologic, cognitive, endocrine and behavioral/psychiatric features.1 It is caused by absent expression of paternally derived genes on the imprinted region at chromosome 15q11.2–q13, and previous studies have indicated that deletions of the paternally derived imprinted region and maternal uniparental disomy 15 (upd(15)mat) account for ∼70 and ∼25% of PWS patients, respectively.1 The remaining PWS patients have rare abnormalities such as epimutations (hypermethylation) of the PWS imprinting center (IC), at the differentially methylated region encompassing exon 1 of SNRPN and microdeletions involving the PWS-IC or HBII-85 small nucleolar RNAs distal to the PWS-IC.2, 3, 4
Upd(15)mat are primarily caused by four mechanisms; that is, trisomy rescue (TR), gamete complementation (GC), monosomy rescue (MR) and post-fertilization mitotic error (PE).5 TR refers to a condition in which chromosome 15 of paternal origin is lost from a zygote with trisomy 15, formed by fertilization between a disomic oocyte and a normal sperm. GC results from fertilization of a disomic oocyte with a nullisomic sperm. MR refers to a condition in which chromosome 15 of maternal origin is replicated in a zygote with monosomy 15, formed by fertilization between a normal oocyte and a nullisomic sperm. PE is an event after formation of a normal zygote. In this regard, a disomic oocyte specific to TR and GC is produced by non-disjunction at meiosis 1 (M1) or meiosis 2 (M2), and non-disjunction at M1 is known to increase with maternal age, probably because of a long-term (10–50 years) meiotic arrest at prophase 1.6
It is predicted, therefore, that the relative frequency of TR/GC-type upd(15)mat through M1 non-disjunction is high in PWS patients born to aged mothers and is increasing in countries where childbearing age is rising. In this context, previous studies have revealed a significantly higher maternal age in PWS patients with upd(15)mat than in those with deletions,7, 8 a significantly higher relative frequency of upd(15)mat in patients born to mothers aged ⩾35 years than in those born to mothers aged <35 years9 and a significantly increased relative frequency of upd(15)mat in PWS patients <5 years of age in United Kingdom where childbearing age is increasing.10 In these studies, however, as underlying mechanisms for upd(15)mat have not been examined, it remains to be clarified whether such maternal age effect on the occurrence of upd(15)mat is primarily mediated by M1 non-disjunction. Furthermore, after studying underlying mechanisms for upd(15)mat by microsatellite analysis, Robinson et al.11 have mentioned that maternal age effect is similar between M1 and M2 errors. Thus, it remains to be clarified whether advanced maternal age is relevant to the occurrence of TR/GC type upd(15)mat through M1 errors.
Here, we report that the advanced maternal age at childbirth constitutes a risk factor for TR/GC type upd(15)mat through M1 non-disjunction.
Materials and methods
This study was approved by the Institute Review Board Committees at the National Center for Child Health and Development and Dokkyo University Koshigaya Hospital, and performed after obtaining informed consent.
PWS patients
This study consisted of 117 Japanese PWS patients (72 male patients and 45 female patients) who satisfied the following selection criteria: (1) normal karyotype in all the 50 lymphocytes examined, (2) hypermethylated PWS-IC that was confirmed by methylation analysis for bisulfite-treated leukocyte genomic DNA, using methylated and unmethylated allele-specific PCR primers (Supplementary Figure 1),12 and (3) positive data on the maternal age at childbirth (parental age was not found in two aged patients who had left our follow-up and whose hospital records had been discarded and in one patient who was born after artificial insemination by donor).
Molecular studies
We performed fluorescence in situ hybridization analysis, microsatellite analysis and multiplex ligation-dependent probe amplification (MLPA) analysis. For fluorescence in situ hybridization analysis, an ∼125-kb probe identifying a region encompassing SNRPN was hybridized to lymphocyte metaphase spreads, together with a CEP 15 probe for D15Z1 and a probe for PML on 15q22 utilized as internal controls. The probe for the SNRPN region was labeled with digoxigenin and detected by rhodamine anti-digoxigenin, and the control probes were detected according to the manufacturer's protocol (Abbott, Chicago, IL, USA). For microsatellite genotyping, PCR amplification was performed for 13 microsatellite loci on chromosome 15, using fluorescently labeled forward primers and unlabeled reverse primers. Subsequently, the PCR products were determined for size on a CEQ8000 autosequencer (Beckman Coulter, Fullerton, CA, USA). For MLPA analysis, we utilized a commercially available MLPA probe mix (ME028-B1) for multiple segments on the chromosome 15 imprinted region, including the PWS-IC and three portions within the HBII-85 small nucleolar RNAs (MRC-Holland, Amsterdam, The Netherlands). The procedure was as described in the manufacturer's instructions. The primers utilized in this study are summarized in Supplementary Table 1.
Classification of PWS patients
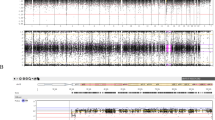
The PWS patients were classified into several groups, according to the underlying (epi) genetic causes (Figure 1). In particular, upd(15)mat was divided into three groups by the previously reported methods13 (Supplementary Figure 2): (1) heterodisomy for at least one of the three adjacent pericentromeric (<4 Mb from the centromere) microsatellite loci (D15S541, D15S542 and D15S1035) was regarded as indicative of TR/GC type upd(15)mat through M1 non-disjunction (TR/GC (M1) group), (2) the combination of isodisomy for the pericentromeric microsatellite loci and heterodisomy for at least one middle to distal microsatellite loci was interpreted as indicative of TR/GC type upd(15)mat through M2 non-disjunction (TR/GC (M2) group) and (3) isodisomy for all the informative microsatellite loci was regarded as indicative of MR/PE type upd(15)mat (MR/PE group). However, it is usually impossible to distinguish between TR and GC, and between MR and PE on the basis of microsatellite data, although identification of segmental isodisomy or mosaicism with a normal cell lineage is unique to PE.14, 15
Analysis of parental ages
We compared parental ages between different groups and between two different time periods (until the year 2002 and since the year 2003), and relative frequency of each group between the two time periods. The setting of the two time periods was based on the Annual Vital Statistics Data from the Japanese Ministry of Health, Labor and Welfare (http://www.mhlw.go.jp/toukei/l ist/81-1.html). The maternal age producing the largest number of live births changed from 25–29 years to 30–34 years, and that producing the third largest number of live births changed from 20–24 years to 35–39 years, between the two time periods (Supplementary Figure 3).
Statistical significance of the median age was examined by the Mann–Whitneys U-test, that of the correlation between parental ages by Spearman's rank correlation test, and that of relative frequency by the Fisher's exact probability test. P<0.05 was considered significant.
Results
Classification of PWS patients
The results are shown in Figure 1. Fluorescence in situ hybridization analysis revealed heterozygous deletions in 84 of the 117 patients (Supplementary Figure 4;Deletion group). Then, microsatellite genotyping was carried out in 27 of the 33 patients without deletions, classifying 15 patients as TR/GC (M1) group, seven patients as TR/GC (M2) group and three patients as MR/PE group (Figure 2;in the remaining six patients, further studies were refused by the parents). There was no finding indicative of segmental isodisomy or mosaicism. Finally, MLPA was performed in the remaining two non-upd(15)mat patients, identifying no microdeletion affecting the PWS-IC. Thus, the two patients were classified as Epimutation group.
Chromosomal locations of the examined microsatellite loci and representative results. MUPD, maternal uniparental disomy (unknown for heterodisomy or isodisomy); mUPD (H), maternal uniparental heterodisomy; mUPD (I), maternal uniparental isodisomy; N.I., not informative. Pericentromeric loci are present in a heterodisomic status in patient 1, and this is consistent with trisomy rescue/gamete complementation (meiosis 1) (TR/GC (M1)) type maternal uniparental disomy 15 (upd(15)mat). For D15S1035, for example, both of the heterozygous maternal alleles are inherited by patient 1, whereas the homozygous paternal alleles are not transmitted to patient 1; this demonstrates mUPD (H) for this locus. In patient 2, all informative loci are present in an isodisomic condition, and this is compatible with monosomy rescue/post-fertilization mitotic error (MR/PE) type upd(15)mat. For D15S1035, for example, one of the two heterozygous maternal alleles is transmitted to patient 2, whereas both of the heterozygous paternal alleles are not inherited by patient 2; this demonstrates mUPD (I) for this locus.
Analysis of parental ages
Distribution of parental ages in each group is shown in Figure 3a, and parental age data are summarized in Table 1. Maternal ages were invariably ⩾35 in TR/GC (M1) group. Furthermore, comparison of maternal ages in Deletion, TR/GC (M1) and TR/GC (M2) groups with >5 patients revealed significant difference between Deletion and TR/GC (M1) groups (P=1.0 × 10−7), but not between Deletion and TR/GC (M2) groups (P=0.19), and between TR/GC (M1) and TR/GC (M2) groups (P=0.085). Paternal ages showed similar tendency, with significant difference between Deletion and TR/GC (M1) groups (P=8.8 × 10−5), but not between Deletion and TR/GC (M2) groups (P=0.39), and between TR/GC (M1) and TR/GC (M2) groups (P=0.39). However, whereas a significant correlation was observed between maternal and paternal ages in Deletion and TR/GC (M2) groups, there was no significant correlation between maternal and paternal ages in TR/GC (M1) group because of relatively advanced maternal ages in this group (Figure 3b). In addition, whereas maternal ages at childbirth were grossly similar between Deletion and TR/GC (M2) groups and the Japanese general population (the mean parental ages at childbirth in Japan were based on the data registered in the Ministry of Health, Labor and Welfare; http://www.mhlw.go.jp/toukei/list/81-1.html), they were obviously higher in TR/GC (M1) group than in the Japanese general population. Paternal ages at childbirth were grossly similar between Deletion group and the Japanese general population and tended to be higher in TR/GC (M1) and TR/GC (M2) groups than in the Japanese general population.
Analysis of parental ages at childbirth. (a) The distribution of parental ages in each group. The light pink and blue vertical bars represent the mean maternal and paternal ages at childbirth from the year 1970 to the year 2008. (b) Correlation between maternal and paternal ages at childbirth. Significant correlation is observed in trisomy rescue/gamete complementation (meiosis 2) (TR/GC (M2)) and Deletion groups, but not in trisomy rescue/gamete complementation (meiosis 1) (TR/GC (M1)) group because of relatively advanced maternal age. (c) Relative frequency of each group in 75 patients born until the year 2002 (n=60, 3, 5, 1, 0 and 6 for Deletion, TR/GC (M1), TR/GC (M2), monosomy rescue/post-fertilization mitotic error (MR/PE), epimutation and non-deletions groups, respectively) and in 42 patients born since the year 2003 (n=24, 12, 2, 2, 2 and 0 for Deletion, TR/GC (M1), TR/GC (M2), MR/PE, Epimutation and Non-deletions groups, respectively).
Relative frequency of each group markedly differed between 75 patients born until 2002 and 42 patients born since 2003 (Figure 3c). Here, TR/GC (M1) was indicated in three of the 75 patients born until the year 2002, and six non-deletion type patients were invariably born until the year 2002. Thus, TR/GC (M1) group accounted for at least three and up to nine of the 75 patients born until the year 2002, and 12 of the 42 patients born since the year 2003. Thus, the relative frequency of TR/GC (M1) was assessed to be significantly different, with the P-values being 1.8 × 10−7 for 3/75 versus 12/42, and 0.025 for 9/75 versus 12/42. In addition, there was no significant change in the parental ages of each group between the two time periods, although the maternal ages at birth of all the patients significantly differed between the two time periods.
Discussion
The present study revealed deletions in 84 patients, upd(15)mat in 25 patients and epimutations in 2 patients. In addition, whereas microsatellite and MLPA analyses were not performed in six patients with non-deletion, the present and the previous studies argue that most of them have upd(15)mat, especially TR/GC (M1) type upd(15)mat.1, 13 Thus, the relative frequency of deletions, upd(15)mat and other rare causes appears to be similar between Japanese patient and previously reported Caucasian patients.1
Notably, the present study implies that advanced maternal age at childbirth constitutes a risk factor for the development of TR/GC (M1) type upd(15)mat. Indeed, maternal ages were significantly higher in TR/GC (M1) group than in Deletion group, which is free from maternal age effect. Although a significant difference was not found between maternal age-dependent TR/GC (M1) group and maternal age-independent TR/GC (M2) group, this would primarily be due to the small number of TR/GC (M2) group. Furthermore, the relative frequency of TR/GC (M1) group significantly increased since the year 2003 when delayed childbearing age became obvious, and the advanced maternal ages at birth since the year 2003 were primarily associated with the high frequency of TR/GC (M1) group rather than the advanced maternal ages in each group. Although it was impossible to distinguish between TR and GC, and between MR and PE,16 this would not pose a major problem. The patients with M1 non-disjunction are included only in TR/GC (M1) group.
Paternal and environmental factors should also be considered for the present results. For a paternal factor, the frequencies of microdeletions and nullisomic sperms might increase with age.17 However, paternal ages at childbirth in each group were similar between the two time periods, and the relative frequency of Deletion group actually decreased since the year 2003. Furthermore, whereas nullisomic sperms can be a background of the development of GC, concomitant occurrence of a nullisomic sperm and a disomic oocyte must be extremely rare. Rather, nullisomic sperms would primarily constitute an underlying factor for the development of maternal age-independent MR. For an environmental factor, it is predicted that chemical materials are increasing with time and that aged parents are exposed to such materials for a long time. In this regard, it has been reported that exposure to environmental chemicals may exaggerate the occurrence of aneuploidies in females.18 Thus, the environmental factor might be relevant to the recent increase of TR/GC (M1) group, although it is unlikely that this factor constitutes the major cause of the increased TR/GC (M1) type upd(15)mat. In males, whereas it has been reported that exposure to chemical materials might facilitate the occurrence of PWS, the relative frequency of genetic causes remained unchanged in PWS patients born to such males.19, 20, 21 Collectively, the effects of such non-maternal age factors would remain small, if any, although further careful examinations are required for the precise evaluation of the maternal age effect on the occurrence of TR/GC (M1).
Several points should be made with regard to the present study. First, we classified upd(15)mat primarily on the basis of the results of three pericentromeric microsatellite loci, with the assumption of no recombination between the centromere and the three loci, as have been employed in the previous study.13 The methods would be basically acceptable, because the three loci reside within a 4 Mb region from the centromere and a recombination is relatively rare in the centromeric regions.22 However, it remains possible that a cryptic recombination(s) might have occurred in the pericentromeric region.
Second, upd(15)mat may also be caused by maternal age-dependent meiotic sister chromatid pre-division that can lead to aneuploid oocytes, including disomic oocytes specific to TR/GC.23 In this regard, as such disomic oocytes can have various patterns of isodisomic and heterodisomic regions, it is impossible to discriminate between upd(15)mat through sister chromatid pre-division and that through conventional meiotic non-disjunction by microsatellite analysis. Thus, the patients classified as TR/GC (M1) group may have upd(15)mat due to maternal age-dependent conventional non-disjunction at M1 and maternal age-dependent sister chromatid pre-division, whereas those classified as TR/GC (M2) group may have upd(15)mat due to maternal age-independent conventional non-disjunction at M2 and maternal age-dependent sister chromatid pre-division. However, even if not all the patients classified as TR/GC (M1) group have upd(15)mat due to conventional non-disjunction at M1, it can be concluded that maternal age-dependent factors still have a critical role in the occurrence of upd(15)mat in patients classified as TR/GC (M1) group. In addition, possible mixture of maternal age-dependent and -independent factors in patients classified as TR/GC (M2) group may be relevant to the lack of significant difference in the maternal age between TR/GC (M2) and Deletion groups, and between TR/GC (M2) and TR/GC (M1) groups.
Lastly, whereas fluorescence in situ hybridization analysis has been routinely performed at commercial laboratories since the year 1993 in Japan, detailed molecular studies including microsatellite analysis are usually available only in institutional laboratories. Thus, a substantial fraction of patients without deletions may have remained undiagnosed or misdiagnosed, without receiving further studies including microsatellite analysis at appropriate institutions. In this regard, considering the opportunity to receive detailed molecular studies, it is possible that upd(15)mat is overlooked more frequently in aged patients than in young patients. If so, this may be relevant to the significant difference in the relative frequency of TR/GC (M1) group between the two time periods (‘since the year 2003’ versus ‘until the year 2002’).
In summary, the results imply that the advanced maternal age at childbirth is a predisposing factor for the development of upd(15)mat because of increased M1 errors. This notion is applicable to maternal upd in general, as well as to trisomies. However, there are several caveats as discussed in the above, and the number of patients, especially those classified as TR/GC (M2) group, is small. Thus, further careful studies using a large number of patients are necessary in the future.
References
Cassidy, S. B. & Driscoll, D. J. Prader-Willi syndrome. Eur. J. Hum. Genet. 17, 3–13 (2009).
Buiting, K., Grob, S., Lich, C., Gillessen-Kaesbach, G., El-Maarri, O. & Horsthemke, B. Epimutation in Prader-Willi and Angelman syndromes: a molecular study of 136 patients with an imprinting defect. Am. J. Hum. Genet. 72, 571–577 (2003).
Sahoo, T., del Gaudio, D., German, J. R., Shinawi, M., Peter, S. U., Person, R. E. et al. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat. Genet. 40, 719–721 (2008).
de Smith, A. J., Purmann, C., Walters, R. G., Ellis, R. J., Holder, S. E., VanHaelst, M. et al. A deletion of the HBII-85 class of small nucleolar RNAs (snoRNAs) is associated with hyperphagia, obesity and hypogonadism. Hum. Mol. Genet. 18, 3257–3265 (2009).
Shaffer, L. G., Agan, N., Goldberg, J. D., Ledbetter, D. H., Longshore, J. W. & Cassidy, S. B. American College of Medical Genetics statement on diagnostic testing for uniparental disomy. Genet. Med. 3, 206–211 (2001).
Jones, K. T. Meiosis in oocytes: predisposition to aneuploidy and its increased incidence with age. Hum. Reprod. Update. 14, 143–158 (2008).
Mitchell, J., Schinzel, A., Langlois, S., Gillessen-Kaesbach, G., Schuffenhauer, S., Michaelis, R. et al. Comparison of phenotype in uniparental disomy and deletion Prader-Willi syndrome: sex specific differences. Am. J. Med. Genet. 65, 133–136 (1996).
Cassidy, S. B., Forsythe, M., Heeger, S., Nicholls, R. D., Schork, N., Benn, P. et al. Comparison of phenotype between patients with Prader-Willi syndrome due to deletion 15q and uniparental disomy 15. Am. J. Med. Genet. 68, 433–440 (1997).
Ginsburg, C., Fokstuen, S. & Schinzel, A. The contribution of uniparental disomy to congenital development defects in children born to mothers at advanced childbearing age. Am. J. Med. Genet. 95, 454–460 (2000).
Whittington, J. E., Butler, J. V. & Holland, A. J. Changing rates of genetic subtypes of Prader-Willi syndrome in the UK. Eur. J. Hum. Genet. 15, 127–130 (2007).
Robinson, W. P., Langlois, S., Schuffenhauer, S., Horsthemke, B., Michaelis, R. C., Christian, S. et al. Cytogenetic and age-dependent risk factors associated with uniparental disomy 15. Prenat. Diagn. 16, 837–844 (1996).
Kubota, T., Das, S., Cristian, S. L., Baylin, S. B., Herman, J. G. & Ledbetter, D. H. Methylation-specific PCR simplifies imprinting analysis. Nat. Genet. 16, 16–17 (1997).
Robinson, W. P., Kuchinka, B. D., Bernasconi, F., Peterson, M. B., Schulze, A., Brondum-Nielsen, K. et al. Maternal meiosis I non-disjunction of chromosome 15: dependence of the maternal age effect on level of recombination. Hum. Mol. Genet. 7, 1011–1019 (1998).
Robinson, W. P. Mechanisms leading to uniparental disomy and their clinical consequences. Bioessays 22, 452–459 (2000).
Kotzot, D. Advanced parental age in maternal uniparental disomy (UPD): implications for the mechanism of formation. Eur. J. Hum. Genet. 12, 343–346 (2004).
Oliver, T. R., Feingold, E., Yu, K., Cheung, V., Tinker, S., Yadav-Shah, M. et al. New insights into human nondisjunction of chromosome 21 in oocytes. PLoS Genet. 4, e1000033 (2008).
Sloter, E., Nath, J., Eskenazi, B. & Wyrobek, A. J. Effects of male age on the frequencies of germinal and heritable chromosomal abnormalities in humans and rodents. Fertil. Steril. 81, 925–943 (2004).
Pacchierotti, F., Adler, I. D., Eichenlaub-Ritter, U. & Mailhes, J. B. Gender effects on the incidence of aneuploidy in mammalian germ cells. Environ. Res. 104, 46–69 (2007).
Strakowski, S. M. & Butler, M. G. Paternal hydrocarbon exposure in Prader-Willi syndrome. Lancet 330, 1458 (1987).
Cassidy, S. B., Gainey, A. J. & Butler, M. G. Occupational hydrocarbon exposure among fathers of Prader-Willi syndrome patients with and without deletions of 15q. Am. J. Hum. Genet. 44, 806–810 (1989).
Akefeldt, A., Anvret, M., Grandell, U., Nordlinder, R. & Gillberg, C. Parental exposure to hydrocarbons in Prader-Willi syndrome. Dev. Med. Chil. Neurol. 37, 1101–1109 (1995).
Robinson, W. P., Bernasconi, F., Mutirangura, A., Ledbetter, D. H., Langlois, S., Malcom, S. et al. Nondisjunction of chromosome 15: origin and recombination. Am. J. Hum. Genet. 53, 740–751 (1993).
Pellestor, F., Andreo, B., Anahory, T. & Hamamah, S. The occurrence of aneuploidy in human: lessons from the cytogenetic studies of human oocytes. Eur. J. Med. Genet. 49, 103–116 (2006).
Acknowledgements
This work was supported by Grants for Research on Intractable Diseases (H22-165) and for Health Research on Children, Youth and Families (H21-005) from the Ministry of Health, Labor and Welfare, and by Grants-in-Aid for Scientific Research (A) (22249010) and Grant-in-Aid for Young Scientists (B) (22791022) from the Japan Society for the Promotion of Science (JSPS).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Matsubara, K., Murakami, N., Nagai, T. et al. Maternal age effect on the development of Prader–Willi syndrome resulting from upd(15)mat through meiosis 1 errors. J Hum Genet 56, 566–571 (2011). https://doi.org/10.1038/jhg.2011.59
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2011.59
Keywords
This article is cited by
-
Risk assessment of assisted reproductive technology and parental age at childbirth for the development of uniparental disomy-mediated imprinting disorders caused by aneuploid gametes
Clinical Epigenetics (2023)
-
Prenatal diagnosis of fetuses with region of homozygosity detected by single nucleotide polymorphism array: a retrospective cohort study
Journal of Human Genetics (2022)
-
Clinical and molecular findings in three Japanese patients with N-acetylneuraminic acid synthetase-congenital disorder of glycosylation (NANS-CDG)
Scientific Reports (2022)
-
Copy neutral absence of heterozygosity on chromosome 15 distal long arm: A surrogate marker for Prader–Willi/Angelman syndromes?
Molecular Cytogenetics (2021)
-
Assisted reproductive technology represents a possible risk factor for development of epimutation-mediated imprinting disorders for mothers aged ≥ 30 years
Clinical Epigenetics (2020)