Abstract
Strong spatial genetic structure in plant populations can increase homozygosity, reducing genetic diversity and adaptive potential. The strength of spatial genetic structure largely depends on rates of seed dispersal and pollen flow. Seeds without dispersal adaptations are likely to be dispersed over short distances within the vicinity of the mother plant, resulting in spatial clustering of related genotypes (fine-scale spatial genetic structure, hereafter spatial genetic structure (SGS)). However, primary seed dispersal by zoochory can promote effective dispersal, increasing the mixing of seeds and influencing SGS within plant populations. In this study, we investigated the effects of seed dispersal by rotational sheep grazing on the strength of SGS and genetic diversity using 11 nuclear microsatellites for 49 populations of the calcareous grassland forb Dianthus carthusianorum. Populations connected by rotational sheep grazing showed significantly weaker SGS and higher genetic diversity than populations in ungrazed grasslands. Independent of grazing treatment, small populations showed significantly stronger SGS and lower genetic diversity than larger populations, likely due to genetic drift. A lack of significant differences in the strength of SGS and genetic diversity between populations that were recently colonized and pre-existing populations suggested that populations colonized after the reintroduction of rotational sheep grazing were likely founded by colonists from diverse source populations. We conclude that dispersal by rotational sheep grazing has the potential to considerably reduce SGS within D. carthusianorum populations. Our study highlights the effectiveness of landscape management by rotational sheep grazing to importantly reduce genetic structure at local scales within restored plant populations.
Similar content being viewed by others
Introduction
Plants due to their sessile life form are highly sensitive to ongoing trends of habitat loss and fragmentation, likely experiencing low rates of immigration of seed and pollen flow within populations from outside sources (Aguilar et al., 2006; Vranckx et al., 2012). As populations become more isolated, spatially restricted pollen flow and seed dispersal within patches lead to an increase of relatedness among neighboring individuals, considerably increasing fine-scale spatial genetic structure (SGS) (Heywood, 1991; Wells and Young, 2002). SGS can increase homozygosity (Ritland, 2002), which reduces the effective size (Ne) of populations, potentially undermining their adaptive genetic potential. Erosion of genetic diversity can affect plant fitness and population viability, increasing the risk of extirpation (Keller and Waller, 2002; Leimu et al., 2006). Evaluating the circumstances that determine the formation of SGS should be an important target in conservation planning of fragmented plant populations. Moreover, spatial patterns of genetic structure are key for understanding ecological and evolutionary dynamics such as changes in reproductive strategies, progeny fitness and local adaptation (Loveless and Hamrick, 1984; Kalisz et al., 2001; Dick et al., 2008; Zhao et al., 2009).
Empirical studies suggest that the strength of SGS is influenced by seed dispersal mode and mating system. Comparisons across species found that species with seed dispersal by gravity show higher SGS than species whose seeds are dispersed by wind or animal vectors (Hardy et al., 2006; Hamrick and Trapnell, 2011; Chung and Chung, 2013), while SGS is usually stronger in selfing relative to outcrossing species (Ennos, 2001; Vekemans and Hardy, 2004). Insights from computer simulations suggest that seed dispersal can have a stronger influence on SGS relative to the dispersal of pollen, but the interaction between selfing rates, pollen and seed dispersal distances, and plant density are critical for the development of SGS (Epperson, 2007). Other factors such as colonization history (Chung et al., 2011; Helsen et al., 2015), demography (Travis et al., 2004; Jacquemyn et al., 2006), or environmental conditions such as habitat fragmentation (Yamagishi et al., 2007; Yue et al., 2012), habitat quality (Bizoux and Mahy, 2007), and land use history (Smith et al., 2009) can also influence the formation of SGS.
In Central Europe, ongoing land use changes had led to a dramatic decline of semi-natural habitats of calcareous grasslands since the 20th century (Poschlod and WallisDeVries, 2002). In particular, the progressive abandonment of rotational grazing by shepherding resulted in numerous extinctions of habitat specialist plants in remnant grasslands due to shrub encroachment and natural reforestation (Butaye et al., 2005). Calcareous grasslands are of high conservation value because they are one of the most species-rich habitats in Central Europe (WallisDeVries et al., 2002). To counteract declines of habitat specialist plants in fragmented grasslands, landscape management practices have been implemented to restore their biodiversity (Aviron et al., 2009). One of such practice relies on the implementation of landscape ecological networks where propagule dispersal can be facilitated through linking grassland patches with rotational grazing by shepherding (Mouissie et al., 2005; Wessels et al., 2008; Wagner et al., 2013). Seed dispersal by domestic ungulates, such as sheep, goats or cattle, has been found to facilitate long-distance dispersal for a range of plant species as herds move along the grazing routes connecting spatially fragmented grasslands (Fischer et al., 1996; Manzano and Malo, 2010; Albert et al., 2015). Because rotational sheep grazing can provide effective seed dispersal and gene flow at the landscape-scale (Rico et al., 2012; Rico et al., 2014b), sheep grazing might also influence the development of fine-scale spatial genetic structure within grazed patches. Experimental studies that have quantified the effects of grazing on the strength of SGS suggest that intensive grazing can negatively impact levels of genetic diversity within populations (Ma et al., 2014; Peng et al., 2015). The effect of grazing on SGS in empirical systems has rarely been studied, and results from available studies are mixed. For instance, grazing has been found to promote aggregation of closely related individuals thus increasing fine-scale genetic structure (Kleijn and Steinger, 2002), while the opposite effect has also been reported (Kloss et al., 2011). Discrepancies between studies are likely related to differences in plant species traits, history of population colonization and land use history.
Landscape management by rotational grazing, which involved the shepherding of large flocks of sheep, has been found to increase plant species colonization rates after local extinctions in previously abandoned grassland patches, irrespective of morphological adaptations to zoochory (Rico et al., 2012; Rico et al., 2014a). Genetic theory predicts that if populations are founded by numerous and unrelated colonists from varied sources, the probability of developing SGS would be low compared with the situation where only a few seed sources provided the initial colonists (Slatkin, 1987; Whitlock and McCauley, 1990). Colonization sets up the primary template of SGS on which seed dispersal and pollen flow will further act (Chung et al., 2003; Trapnell et al., 2013). Moreover, colonization success of restored habitats for establishing long-term persistent plant populations might largely depend on their initial genetic make up such as degree of genetic diversity and SGS (Whitlock and McCauley, 1990; Mijangos et al., 2015).
In this study, we investigated the effect of rotational sheep grazing on the strength of SGS within plant populations of recently restored calcareous grasslands in southern Germany. We studied the perennial herb and specialist of calcareous grasslands Dianthus carthusianorum (Oberdorfer, 1978), which lacks seed-dispersal traits related to zoochory or anemochory, and does not reproduce vegetatively (Klotz et al., 2002). This landscape-scale network of calcareous grassland patches is an ideal empirical system because it allows us to investigate the intensity of SGS across multiple plant populations within the same landscape and with known management records for all analyzed patches, which previously has not been done. Evaluating if landscape management by rotational sheep grazing can modify the development of SGS in previously fragmented plant populations is relevant to better inform conservation planning. Previous work in the study area for D. carthusianorum found that gene flow at the landscape scale and genetic diversity within populations were associated with connectivity facilitated by rotational sheep grazing (Rico et al., 2014a,b). Here, we therefore ask the following: (i) does rotational sheep grazing have a significant effect on patterns of SGS at the local scale, within grassland patches? (ii) Does this effect depend on population size? For grazed grasslands, we further compare SGS and genetic diversity between recently colonized patches (since 1989) and populations existing before the implementation of the ecological network in our study area. Specifically, we ask (iii) do colonized populations have lower genetic diversity than pre-existing ones, indicating a recent founder event? (iv) Do recently colonized populations exhibit stronger patterns of SGS relative to pre-existing populations?
Materials and methods
Study landscape and species
The study was conducted in the Southern Franconian Alb near Weissenburg, Bavaria, Germany covering an area of ~10 × 15 km. Calcareous grasslands of the Gentiano-Koelerietum pyramidatae vegetation association (Oberdorfer, 1978) are mainly located on the steep slopes between the Upper Franconia Jura plateau and the valleys. In our study area, management of calcareous grasslands through rotational sheep grazing was initiated in 1989 to reconnect grassland patches that had been abandoned at least since 1960 with grasslands where grazing had been continued throughout the 20th century (core areas; Wagner et al., 2013). Flocks of 400–800 ewes with their lambs are herded along defined routes in three non-overlapping shepherding systems. Since the start of the project in 1989, out of 62 previously abandoned grassland patches, 26 were subjected to grazing three to five times annually throughout the season, 13 were grazed only toward the end of the season or only during a few years at the beginning of the project, whereas 23 remained ungrazed (Rico et al., 2012). In the study area, sheep are kept in designated paddocks for rumination as prescribed by calcareous grassland conservation management. Consumed seeds are thus unlikely to be deposited in grassland patches, so that dispersal might depend mostly on epizoochory (Rico et al., 2014b).
D. carthusianorum (Caryophyllaceae) is a perennial herb of 30–45 cm in height. It is predominantly outcrossing and does not form a persistent seed bank (Bloch et al., 2005). According to Hensen (1997), D. carthusianorum reproduces sexually, and its clonal growth is of the ‘phalanx’ type that lacks lateral spread. Klotz et al. (2002) specify the clonal growth form as ‘rhizome-like pleiocorm’, where adventitiously rooted shoots may lose their connection with the primary roots. Flowering occurs from June to October and pollination is carried out by specialized Lepidoptera species (Bloch et al., 2005). The seeds lack morphological adaptations to dispersal by wind or animals (Klotz et al., 2002).
Sampling and laboratory procedures
Detailed sampling information and microsatellite genotype data at 11 polymorphic loci are described in Rico et al., 2014a,b (available on DRYAD doi:10.5061/dryad.jn1v7). In brief, we collected leaf material from all adult plants in patches with less than 40 individuals (small populations), whereas 30–40 adult plants were sampled from patches with more than 40 individuals (large populations). The latter implied that sampling was spread out within patches of large populations. During the field sampling collection, we did not observe differences in the density or spatial aggregation of individuals between small and large populations. To maintain comparability, we excluded patches with <10 individuals, one patch with a minimum distance between neighboring individuals >10 m, and four patches that were grazed only during a few years but remained ungrazed for the last 15 years. Geographic coordinates of each sampled individual were recorded using a Trimble GeoXT 2008 (Westminster, CO, USA) GPS receiver with sub-meter resolution based on differential GPS post-processing. The final data set included the genotypes of 1304 adult individuals of 49 patches (referred here as populations; Figure 1).
Schematic representation of the spatial configuration of patches analyzed for SGS in D. carthusianorum. Black circles denote larger populations and white circles correspond to small populations (<40 individuals). Dashed lines indicate rotational sheep grazing routes connecting calcareous grassland patches in three non-overlapping grazing systems. Ungrazed populations are showed as white (small populations) or black (large populations) squares and without connecting lines. Gray shaded areas in the background show forested areas. The inset map shows the distribution of calcareous grasslands (gray areas) in Germany and the location of the study area (map modified from Beinlich and Plachter, 1995).
Quantification of SGS and genetic diversity
To evaluate the patterns of SGS, we calculated a genetic distance matrix using the multilocus measure of the pairwise kinship coefficient Fij of Ritland (1996), which included all individual pairwise comparisons in the whole-data-set without specifying spatial distance classes or groups, as implemented in SPAGEDI (Hardy and Vekemans, 2002). The Fij coefficient measures the probability that two alleles are identical by descent. Negative values of Fij can occur if allele frequencies between two compared individuals differ more than expected at random from the entire data set. To assess the strength of SGS within each patch, we estimated the Sp statistic of Vekemans and Hardy (2004). The Sp statistic has been widely applied in empirical studies of SGS since it fits a linear model that estimates SGS under isolation by distance. The Sp statistic is very robust to sampling scheme and definitions of distance classes, which allows comparisons among plots and among species (Vekemans and Hardy, 2004). If Fij decrease linearly with the logarithm of distance, the strength of SGS can be quantified by the Sp statistic as bF/(1-F1), where bF represents the regression slope of the pairwise Fij relatedness coefficients and F1 is the mean Fij between pairs of individuals at the shortest distance class (Vekemans and Hardy, 2004). Since we have differences in the sampling of individuals between small and large populations, distance classes were not defined per patch, instead F1 was defined as the regression intercept, that is, the value where the logarithm of distance is 0, which corresponds to the value at a distance of 1 m. Here, F1 represents the expected degree of the relatedness coefficient between individuals 1 m distance apart. Higher values of the Sp statistic indicate a high rate of distance-decay of kinship. Significance of the regression slope in each population was tested by permuting the pairwise Fij coefficients within populations 1000 times. Analyses were implemented using R (R CoreTeam, 2014). Genetic diversity indices including allelic richness (Ar), observed (Ho) and unbiased expected heterozygosity (He) were estimated for each population using GENALEX (Peakall and Smouse, 2012). We corrected the measures of allelic richness for the effect of sample size using the rarefaction procedure in HP-RARE (Kalinowski, 2005). FST indices of genetic differentiation among populations within patches of the same type (for example, colonized vs pre-existing) and their s.d. were estimated by jackknifing loci over populations using SPAGEDI (Hardy and Vekemans, 2002).
Statistical analysis
We used a two-way analysis of variance (ANOVA) to test for main effects and interaction of the two factors: grazing (two classes: grazed and ungrazed) and population size (two ordinal classes: <40 individuals vs ⩾40 individuals) on each of five response variables: the three genetic diversity indices (Ar, Ho and He) and the F1 and Sp statistics. To account for an unbalanced design, we used type II sums of squares. To normalize the distribution of residuals, we applied square-root transformation to F1 and Sp statistic, while genetic diversity indices were not transformed. Negative values of the Sp statistic were set to zero. Model assumptions of normal distribution of residuals and homogeneity of variances were visually checked. In addition, we tested if there were differences in the degree of spatial isolation between ungrazed vs grazed patches using the Welch two-sample t-test of a Si patch connectivity index estimated in Rico et al. (2012).
Since population history (that is, presence of D. carthusianorum before reintroduction of sheepherding 1989) is not confirmed for all core patches, statistical analysis of population history was only performed for populations with known history: populations present at 1989 survey (pre-existing populations, n=7) and new populations (colonized patches, n=9) which were absent in 1989 but present in the 2009 survey. Owing to reduced sample size, population history was analyzed separately but not included as an additional factor in the two-way ANOVA with the factors grazing and population size. All statistical analyses were performed in R (R CoreTeam, 2014).
Results
Strength of SGS
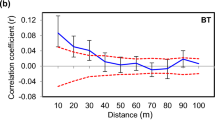
SGS in D. carthusianorum was significant in 26 out of 49 populations (Table 1). The pairwise kinship coefficient Fij showed a distance decay in all treatment groups (Supplementary Figure S1). The strength of SGS as quantified by the Sp statistic was significantly related to both grazing and population size, and without a significant interaction (two-way ANOVA: sheep grazing: F1,45=4.74, P=0.035; population size: F1,45=4.28, P=0.044; and interaction: F1,45=0.003, P=0.954). Grazed populations showed significantly weaker SGS than ungrazed populations (that is, lower Sp values), and independent of grazing, small populations showed significantly higher Sp values than large populations (Figure 2). Moreover, the regression slopes bF of all ungrazed patches, independent of population size, were significant, while for grazed grasslands, small populations showed a higher number of significant cases of SGS than large populations (small: 6 out of 11, large: 13 out of 32 populations; Table 1).
Interaction plot of the mean square-root transformed Sp statistic as a function of the factors grazing and population size in populations of D. carthusianorum. Both factors were statistically different and without an interaction. The dashed line indicates small population size (F1,45=4.74, P=0.035) and the full-line large population size (F1,45=4.28, P=0.044).
There were marginally significant effects of grazing (F1,45=3.47, P=0.068) and no significant effects on population size (F1,45=1.87, P=0.17) on F1, without significant interaction (F1,45=0.15, P=0.701). F1 values varied across populations (Table 2) and overall, F1 mean values were small for all treatment groups (large grazed: F1=0.027, small grazed: F1=0.040, small ungrazed: F1=0.074, large ungrazed: F1=0.048), which indicated that the nearest neighbors (at 1 m distance) were not highly related. On the other hand, there were no significant differences in the degree of spatial isolation between populations of ungrazed and grazed grassland patches (df=10.7, t=1.61 and P=0.13). None of the models showed unusual patterns in the residuals distribution or influential outliers.
Within grazed grasslands, the strength of SGS (Sp statistic) was higher on average in pre-existing populations relative to colonized populations (Figure 3), but the difference was not statistically significant (ANOVA, F1,14=0.7, P=0.416). The difference remained non-significant when accounting for population size (two-way ANOVA: population history F1,12=4.03, P=0.067; population size: F1,12=2.7, P=0.127; interaction: F1,12=1.26, P=0.282). Similarly, F1 values were not statistically significantly different between pre-existing and colonized populations (ANOVA, F1,14=3.09, P=0.587).
Estimates of genetic diversity
Average estimates of genetic diversity tended to be higher in large and small populations of grazed patches relative to large and small populations of ungrazed grasslands (except for Ho in ungrazed-small vs grazed-small), with the largest difference for mean allelic richness (Table 2). ANOVA showed that both sheep grazing and population size had a significant effect only on mean allelic richness, and without significant interaction between factors (two-way ANOVA: sheep grazing: F1,44=7.09, P=0.011; population size: F1,44=6.3, P=0.016; and interaction: F1,44=1.28, P=0.263). No significant differences were observed for Ho (two-way ANOVA: sheep grazing: F1,45=0.48, P=0.492; and population size: F1,45=3.2, P=0.081) and neither for He (two-way ANOVA: sheep grazing: F1,45=0.08, P=0.772; and population size: F1,45=0.36, P=0.546). The FST index was highest for small, ungrazed populations (FST=0.054), whereas the lowest value was for large, grazed populations (FST=0.018).
Colonized populations had similar values of Ho (0.53), He (0.59) and mean Ar (4.3) than pre-existing populations (Ho=0.54, He=0.53 and Ar=4.4), and thus no significant differences were found (ANOVA: Ar: df=2, F=0.98, P=0.395; Ho: df=2, F=1.2, P=0.326; and He: df=2, F=2.23, P=0.139). The FST was higher in colonized populations relative to pre-existing populations, but this difference was not large (Table 3).
Discussion
We tested, within an empirical landscape system, the effects of rotational sheep grazing on the strength of SGS across multiple populations of a habitat specialist plant of calcareous grasslands. Our results showed that D. carthusianorum populations under rotational sheep grazing showed significantly weaker SGS than populations of ungrazed grasslands. Moreover, our results showed that recently colonized populations of grazed grasslands did not show stronger patterns of SGS nor lower genetic diversity than pre-existing populations of grazed grasslands. Together these results suggest that rotational sheep grazing not only increases landscape-scale genetic connectivity of D. carthusianorum among patches (Rico et al., 2014b) but also plays an important role in reducing SGS in previously abandoned calcareous grasslands, likely by increasing the overlap of seed shadows. However, small plant populations showed significantly stronger SGS relative to large populations, and this effect was independent of grazing.
SGS and grazing effects
Our results are consistent with previous empirical evidence showing that species dispersed by zoochory (but mostly frugivores) exhibit weak SGS, which might be explained by effective seed dispersal over larger distances (Degen et al., 2001; Hamrick and Trapnell, 2011; Wang et al., 2011). Rico et al. (2014b) showed that rotational sheep grazing is an important determinant of spatial genetic structure at the landscape scale in D. carthusianorum, which was associated with long-distance seed dispersal among populations connected through the grazing route. Here, we demonstrated that rotational sheep grazing can as well influence spatial patterns of fine-scale genetic structure, even for a species lacking morphological adaptations to zoochory.
Few empirical studies have investigated the effects of grazing on genetic diversity and genetic structure at local spatial scales. For Veronica chamaedrys, Kloss et al. (2011) found that genetic structure was influenced by land use type, where populations in grazed grasslands showed lower genetic structure compared with mown pastures and meadows, but no effects on genetic diversity. For Veratrum album, Kleijn and Steinger (2002) also did not find any effect of grazing on genetic diversity within populations, but found that grazing increased the spatial aggregation of related genotypes relative to the strength of SGS of plant populations in meadows. The significant pattern of SGS in V. album for grazed grasslands was related to an enhancement of clonal growth by grazing and lower seedling recruitment (Kleijn and Steinger, 2002). In contrast to these studies, our results for D. carthusianorum indicate that landscape management by rotational sheep grazing influenced both genetic diversity and fine-scale genetic structure in small and large plant populations of previously abandoned grasslands. Formation of SGS in D. carthusianorum cannot be explained by vegetative propagation, as duplicated genotypes were practically absent (approximately <10 individuals). Moreover, the Sp values in D. carthusianorum fall within the range of values expected for outcrossing species: observed values ranged from 0.005 to 0.022, similar to Sp values expected for outcrossing species (Sp: 0.013), relative to higher values expected for mixed mating systems (Sp: 0.037; Vekemans and Hardy, 2004).
We did not observe significant effects on F1 values for grazing or population size, although both small and large populations of ungrazed grasslands showed higher F1 values relative to grazed grasslands. Significant differences in Sp but not in F1 (mean Fij value at 1 m distance) between grazed and ungrazed patches, might indicate that patterns of SGS in D. carthusianorum are largely determined by seed dispersal, while pollen-mediated gene flow is likely less restricted (Sork, 2016). While all populations of ungrazed patches showed significant patterns of SGS, only 42% of populations in grazed patches showed significant SGS. There are two likely mechanisms by which grazing can reduce patterns of SGS: seed dispersal (Fischer et al., 1996; Manzano and Malo, 2010) and small-scale disturbances (Adler et al., 2001; Faust et al., 2011). For instance, local disturbances from grazing can reduce SGS by trampling and grubbing (Adler et al., 2001) creating thinning of vegetation that promotes recruitment (Chung et al., 2003). In the case of dispersal, sheep can increase the variance of dispersal distances by attachment to the fur or hooves of sheep (Couvreur et al., 2004). This will lead to a greater overlap of seed shadows from different plants within fragments compared with ungrazed grasslands (Hamrick and Trapnell, 2011).
SGS and population size effects
Population size is another important factor modifying SGS within populations. We observed that independent of grazing, small populations (<40 individuals) had significantly stronger SGS than large populations. For instance, within grazed grasslands, small populations exhibited stronger SGS and had higher proportions of cases (55%) of significant SGS than large grazed populations (40%). In small populations, which experience high rates of genetic drift, development of SGS can be intensified due to increased mating between relatives thus reducing Ne, which has been observed in small isolated populations in fragmented landscapes (for example, O’Connell et al., 2006; Mimura and Aitken, 2007). Low plant density, which in turn could lead to a reduced overlap of seed shadows, is expected to influence the development of SGS (Hamrick and Trapnell, 2011). However, recent evidence showed that individual spatial aggregation instead of plant density is more relevant for the development of SGS (Lara-Romero et al., 2016). We do not have data on plant density to analyze any potential effects on SGS in small vs large populations. Here, we observed that small populations of grazed grasslands had significantly lower SGS and higher genetic diversity than small populations in ungrazed grasslands. Previous evidence showed that mean allelic richness in D. carthusianorum populations was positively correlated with patch connectivity through rotational sheepherding, where ungrazed patches had lower allelic richness (Rico et al., 2014b). The increase of seed mixing from varied sources facilitated by rotational sheep grazing can influence levels of genetic diversity within populations, and this effect could be expected more relevant for small populations than spatial isolation per se. Moreover, FST estimates of genetic differentiation were the highest among small, ungrazed grasslands. Overall our results suggest that gene flow was less restricted among grazed grasslands, which in turn would increase the effective size of populations (Ne).
SGS and population history
In accordance with our expectations, we did not find differences in the strength of SGS and genetic diversity between recently colonized and pre-existing populations of grazed patches. Restoration of calcareous grasslands by rotational shepherding since 1989 promoted colonization by diverse plant species in numerous grasslands patches (Rico et al., 2012). Before the implementation of the landscape management project, D. carthusianorum occurred in 44% of 62 previously abandoned patches and in more than 90% of the core areas (Boehmer et al., 1990). An evaluation survey in 2009 showed that successful colonizations increased the species’ occurrence to 82% in previously abandoned patches, whereas occurrence in ungrazed patches remained at 38%. Our results provide insights in the colonization process of previously abandoned grasslands after reintroduction of rotational sheepherding in the study area. Theoretical genetic models of colonization predict that genetic diversity will be low and genetic differentiation among recently colonized populations will be high if populations are founded by colonists from few sources (propagule pool model). On the other hand, high genetic diversity and low genetic differentiation will occur if colonists are from diverse seed sources (migrant pool model; Whitlock and McCauley, 1990; Pannell and Dorken, 2006). The lack of significant differences in SGS, values of genetic diversity and FST estimates of genetic differentiation between recently colonized and pre-existing populations in grazed grasslands, suggest that population founders likely came from varied sources. A migrant pool model of colonization likely associated with seed dispersal by sheep has also been reported for the calcareous grassland forb Anthyllis vulneraria (Helsen et al., 2015). Empirical studies in expanding populations have found that the strength of SGS can decrease as populations mature due to seedling mortality during the establishment process, and by competition within individuals of the same cohort (Chung et al., 2003; Tero et al., 2005). However, the opposite has also been reported as the result of restricted seed dispersal and aggregation of juveniles around the mother plant (Chung et al., 2011). Unfortunately, we were unable to test the effect of population size on population history because most of colonized populations were small, while many pre-existing populations were large.
Spatially restricted gene flow at the landscape and local scales in outcrossing plant species can negatively increase homozygosity, which in turn affects individual fitness and the viability of populations in the long run (Leblois et al., 2006). These effects are predicted to become stronger as populations become smaller (Keller and Waller, 2002). Although we observed low levels of inbreeding in D. carthusianorum, small populations exhibited higher inbreeding relative to large populations (Rico et al., 2014a). Small populations should be important targets in conservation management as high rates of genetic drift can considerably compromise the adaptive potential of populations to persist under ongoing climatic changes (Eizaguirre and Baltazar-Soares, 2014). Our results have important implications for landscape management and restoration as we found a positive effect of rotational shepherding in terms of a significant reduction of SGS across plant populations in previously abandoned grasslands. Importantly, this effect is more relevant for small populations because a decrease of SGS would tend to increase Ne and in turn reduce the negative effects of genetic drift on levels of genetic diversity. Our study highlights the effectiveness of landscape management by rotational sheep grazing to modify spatial patterns of genetic structure at local scales within plant populations of restored grasslands. Further investigations should establish whether the observed effects of grazing on fine-scale SGS are consistent across a range of typical grassland species.
DATA ARCHIVING
Population and individual microsatellite genotype data available on Dryad doi:10.5061/dryad.jn1v7.
References
Adler P, Raff D, Lauenroth W . (2001). The effect of grazing on the spatial heterogeneity of vegetation. Oecologia 128: 465–479.
Aguilar R, Ashworth L, Galetto L, Aizen MA . (2006). Plant reproductive susceptibility to habitat fragmentation: review and synthesis through a meta-analysis. Ecol Lett 9: 968–980.
Albert A, Auffret AG, Cosyns E, Cousins SAO, D’hondt B, Eichberg C et al. (2015). Seed dispersal by ungulates as an ecological filter: a trait-based meta-analysis. Oikos 124: 1109–1120.
Aviron S, Nitsch H, Jeanneret P, Buholzer S, Luka H, Pfiffner L et al. (2009). Ecological cross compliance promotes farmland biodiversity in Switzerland. Front Ecol Environ 7: 247–252.
Beinlich B, Plachter H . (1995). Ein Naturschutzkonzept für die Kalkmagerrasen der Mittleren Schwäbischen Alb (Baden-Württemberg): Schutz, Nutzung und Entwicklung. Beihefte zu den Veröffentlichungen für Naturschutz und Landschaftspflege in Baden-Württemberg 83, pp 1–520.
Bizoux JP, Mahy G . (2007). Within-population genetic structure and clonal diversity of a threatened endemic metallophyte, Viola calaminaria (Violaceae). Am J Bot 94: 887–895.
Bloch D, Werdenberg N, Erhardt A . (2005). Pollination crisis in the butterfly-pollinated wild carnation Dianthus carthusianorum? New Phytol 169: 699–706.
Boehmer HJ, Janeck L, Steidler S, Raab B . (1990) Verbundsystem Halbtrockenrasen. Trittstein- und Refugialbiotope im östlichen Landkreis Weißenburg-Gunzenhausen-Technical report, Bayerisches Landesamt für Umweltschutz, LfU, Munich, Germany.
Butaye J, Adriaens D, Honnay O . (2005). Conservation and restoration of calcareous grasslands: a concise review of the effects of fragmentation and management on plant species. Biotechnol Agron Soci Environ 9: 111–118.
Couvreur M, Vandenberghe B, Verheyen K, Hermy M . (2004). An experimental assessment of seed adhesivity on animal furs. Seed Sci Res 14: 147–159.
Chung MY, Chung MG . (2013). Significant spatial aggregation and fine-scale genetic structure in the homosporous fern Cyrtomium falcatum (Dryopteridaceae). New Phytol 199: 663–672.
Chung MY, Epperson BK, Chung MG . (2003). Genetic structure of age classes in Camellia japonica (Theaceae). Evolution 57: 62–73.
Chung MY, Nason JD, Chung MG . (2011). Significant demographic and fine-scale genetic structure in expanding and senescing populations of the terrestrial orchid Cymbidium goeringii (Orchidaceae). Am J Bot 98: 2027–2039.
Degen B, Caron H, Bandou E, Maggia L, Chevallier MH, Leveau A et al. (2001). Fine-scale spatial genetic structure of eight tropical tree species as analysed by RAPDs. Heredity 87: 497–507.
Dick CW, Hardy OJ, Jones FA, Petit RJ . (2008). Spatial scales of pollen and seed-mediated gene flow in tropical rain forest trees. Trop Plant Biol 1: 20–33.
Eizaguirre C, Baltazar-Soares M . (2014). Evolutionary conservation-evaluating the adaptive potential of species. Evol Appl 7: 963–967.
Ennos RA : (2001). Inferences about spatial processes in plant populations from the analysis of molecular markers. In: Silvertown J, Antonovics J (eds) Integrating Ecology And Evolution In A Spatial Context. Blackwell Science. pp 45–57.
Epperson BK : (2007). Plant dispersal, neighborhood size and isolation by distance. Mol Ecol 16: 3854–3865.
Faust C, Eichberg C, Storm C, Schwabe A . (2011). Post-dispersal impact on seed fate by livestock trampling - A gap of knowledge. Basic Appl Ecol 12: 215–226.
Fischer SF, Poschlod P, Beinlich B . (1996). Experimental studies on the dispersal of plants and animals on sheep in calcareous grasslands. J Appl Ecol 33: 1206–1222.
Hamrick JL, Trapnell DW . (2011). Using population genetic analyses to understand seed dispersal patterns. Acta Oecologica 37: 641–649.
Hardy OJ, Maggia L, Bandou E, Breyne P, Caron H, Chevallier M-H et al. (2006). Fine-scale genetic structure and gene dispersal inferences in 10 neotropical tree species. Mol Ecol 15: 559–571.
Hardy OJ, Vekemans X . (2002). spagedi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2: 618–620.
Helsen K, Jacquemyn H, Honnay O . (2015). Hidden founder effects: small-scale spatial genetic structure in recently established populations of the grassland specialist plant Anthyllis vulneraria. Mol Ecol 24: 2715–2728.
Hensen I . (1997). Life strategy systems of xerothermic grasslands - mechanisms of reproduction and colonization within Stipetum capillatae s.1. and Adonido - Brachypodietum pinnati. Feddes Repert 108: 425–452.
Heywood J . (1991). Spatial analysis of genetic variation in plant populations. Annu Rev Ecol Syst 22: 335–355.
Jacquemyn H, Brys R, Vandepitte K, Honnay O, Roldán-Ruiz I . (2006). Fine-scale genetic structure of life history stages in the food-deceptive orchid Orchis purpurea. Mol Ecol 15: 2801–2808.
Kalinowski ST . (2005). HP-RARE 1.0: A computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5: 187–189.
Kalisz S, Nason JD, Hanzawa FM, Tonsor SJ . (2001). Spatial population genetic structure in Trillium grandiflorum: the roles of dispersal, mating, history, and selection. Evolution 55: 1560–1568.
Keller LF, Waller DM . (2002). Inbreeding effects in wild populations. Trends Ecol Evol 17: 230–241.
Kleijn D, Steinger T . (2002). Contrasting effects of grazing and hay cutting on the spatial and genetic population structure of Veratrum album, an unpalatable, long-lived, clonal plant species. J Ecol 90: 360–370.
Kloss L, Fischer M, Durka W . (2011). Land-use effects on genetic structure of a common grassland herb: A matter of scale. Basic Appl Ecol 12: 440–448.
Klotz S, Ingolf K, Durka W . (2002) BIOLFLOR: eine Datenbank mit biologisch-Ökologischen Merkmalen zur Flora von Deutschland. Bundesamt fur Naturschutz: Bonn, Germany.
Lara-Romero C, Garcia-Fernandez A, Robledo-Arnuncio JJ, Roumet M, Morente-Lopez, Lopez-Gil A et al. (2016). Individual spatial aggregation correlates with between-population variation in fine-scale genetic structure of Silene ciliata (Caryophyllaceae). Heredity 116: 417–423.
Leblois R, Estoup A, Streiff R . (2006). Genetics of recent habitat contraction and reduction in population size: does isolation by distance matter? Mol Ecol 15: 3601–3615.
Leimu R, Mutikainen P, Koricheva J, Fischer M . (2006). How general are positive relationships between plant population size, fitness and genetic variation? J Ecol 94: 942–952.
Loveless MD, Hamrick JL . (1984). Ecological determinants of genetic structure in plant populations. Annu Rev Ecol Syst 15: 65–95.
Ma DT, Guo YX, Hou FJ, Zhai XY, Wang W, Tian M et al. (2014). Plant genetic diversity and grazing management on the Qinghai-Tibetan Plateau: a case study of a dominant native wheatgrass (Elymus nutans). Biochem Syst Ecol 56: 16–23.
Manzano P, Malo JE . (2010). Extreme sheep long-distance seed dispersal via sheep. Front Ecol Environ 4: 244–248.
Mijangos JL, Pacioni C, Spencer PBS, Craig MD . (2015). Contribution of genetics to ecological restoration. Mol Ecol 24: 22–37.
Mimura M, Aitken SN . (2007). Increased selfing and decreased effective pollen donor number in peripheral relative to central populations in Picea sitchensis (Pinaceae). Am J Bot 94: 991–998.
Mouissie AM, Vos P, Verhagen HMC, Bakker JP . (2005). Endozoochory by free-ranging, large herbivores: ecological correlates and perspectives for restoration. Basic Appl Ecol 6: 547–558.
O’Connell LM, Mosseler A, Rajora OP . (2006). Impacts of forest fragmentation on the mating system and genetic diversity of white spruce (Picea glauca at the landscape level. Heredity 97: 418–426.
Oberdorfer E . (1978) SÜddeutsche Pflanzengesellschaften vol. 2, Gustav Fischer Verlag: Stuttgart, Germany.
Pannell JR, Dorken ME . (2006). Colonisation as a common denominator in plant metapopulations and range expansions: effects on genetic diversity and sexual systems. Landsc Ecol 21: 837–848.
Peakall R, Smouse PE . (2012). GenALEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28: 2537–2539.
Peng J, Liang C, Niu Y, Jiang W, Wang W, Wang L . (2015). Moderate grazing promotes genetic diversity of Stipa species in the Inner Mongolian steppe. Landsc Ecol 30: 1783–1794.
Poschlod P, WallisDeVries MF . (2002). The historical and socioeconomic perspective of calcareous grasslands - Lessons from the distant and recent past. Biol Conserv 104: 361–376.
R CoreTeam. (2014) R: a Language and Environment for Statistical Computing R Foundation for Statistical Computing: Vienna, Austria.
Rico Y, Boehmer HJ, Wagner HH . (2012). Determinants of actual functional connectivity for calcareous grassland communities linked by rotational sheep grazing. Landsc Ecol 27: 199–209.
Rico Y, Boehmer J, Wagner HH . (2014a). Effect of rotational shepherding on demographic and genetic connectivity of calcareous grassland plants. Conserv Biol 28: 467–477.
Rico Y, Holderegger R, Boehmer HJ, Wagner HH . (2014b). Directed dispersal by rotational shepherding supports landscape genetic connectivity in a calcareous grassland plant. Mol Ecol 23: 832–842.
Ritland K . (1996). Estimators for pairwise relatedness and individual inbreeding coefficients. Genet Res 67: 175–185.
Ritland K . (2002). Extensions of models for the estimation of mating systems using n independent loci. Heredity 88: 221–228.
Slatkin M . (1987). Gene flow and the geographic structure of natural populations. Science 236: 787–792.
Smith SE, Arredondo T, Aguiar M, Huber-Sannwald E, Alpuche A, Aguado A et al. (2009). Fine-scale spatialg enetic structure in perennial grasses in three Environments. Rangel Ecol Manag 62: 356–363.
Sork VL . (2016). Gene flow and natural selection shape spatial patterns of genes in tree populations: Implications for evolutionary processes and applications. Evol Appl 9: 291–310.
Tero N, Aspi J, Siikamäki P, Jäkäläniemi A . (2005). Local genetic population structure in an endangered plant species, Silene tatarica (Caryophyllaceae). Heredity 94: 478–487.
Trapnell DW, Hamrick JL, Ishibashi CD, Kartzinel TR . (2013). Genetic inference of epiphytic orchid colonization; it may only take one. Mol Ecol 22: 3680–3692.
Travis SE, Proffitt CE, Ritland K . (2004). Population structure and inbreeding vary with successional stage in created Spartina alterniflora marshes. Ecol Appl 14: 1189–1202.
Vekemans X, Hardy OJ . (2004). New insights from fine-scale spatial genetic structure analyses in plant populations. Mol Ecol 13: 921–935.
Vranckx G, Jacquemyn H, Muys B, Honnay O . (2012). Meta-analysis of susceptibility of woody plants to loss of genetic diversity throughhabitat fragmentation. Conserv Biol 26: 228–237.
Wagner HH, Rico Y, Lehnert H, Boehmer HJ . (2013). Process-based long-term evaluation of an ecological network of calcareous grasslands connected by sheep herding. Ecography 36: 374–382.
WallisDeVries MF, Poschlod P, Willems JH . (2002). Challenges for the conservation of calcareous grasslands in northwestern Europe: integrating the requirements of flora and fauna. Biol Conserv 104: 265–273.
Wang R, Compton SG, Chen X-Y . (2011). Fragmentation can increase spatial genetic structure without decreasing pollen-mediated gene flow in a wind-pollinated tree. Mol Ecol 20: 4421–4432.
Wells GP, Young AG . (2002). Effects of seed dispersal on spatial genetic structure in populations of Rutidosis leptorrhychoides with different levels of correlated paternity. Genet Res 79: 219–226.
Wessels S, Eichberg C, Storm C, Schwabe A . (2008). Do plant-community-based grazing regimes lead to epizoochorous dispersal of high proportions of target species? Flora - Morphol Distrib Funct Ecol Plants 203: 304–326.
Whitlock MC, McCauley DE . (1990). Some population genetic consequences of colony formation and extinction: genetic correlations within founding groups. Evolution 44: 1717–1724.
Yamagishi H, Tomimatsu H, Ohara M . (2007). Fine-scale spatial genetic structure within continuous and fragmented populations of Trillium camschatcense. J Hered 98: 367–372.
Yue X-L, Chen J-M, Guo Y-H, Wang Q-F . (2012). Fine-scale spatial genetic structure of an endangered marsh herb, Caldesia grandis (Alismataceae). Genet Mol Res 11: 2412–2421.
Zhao R, Xia H, Lu BR . (2009). Fine-scale genetic structure enhances biparental inbreeding by promoting mating events between more related individuals in wild soybean (Glycine soja; Fabaceae) populations. Am J Bot 96: 1138–1147.
Acknowledgements
This research was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC Discovery Grant to HHW), the National Council on Science and Technology of Mexico (CONACYT # 303735 to YR), and the German Academic Exchange Service (DAAD to YR). We thank HJ Boehmer, K Dadrich, D Baumgartner, B Raab, S Haacke, J Sachteleben and the shepherds E Beil, E Neulinger and A Grimm for valuable information, and Henry Lehnert for assistance during sample collection.
Author contributions
YR: conception and design of research, field work, molecular genetic lab work, statistical analysis, main writing of paper and editing. HHW: conception and design of research, field work, statistical analysis and editing of manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Heredity website
Supplementary information
Rights and permissions
About this article
Cite this article
Rico, Y., Wagner, H. Reduced fine-scale spatial genetic structure in grazed populations of Dianthus carthusianorum. Heredity 117, 367–374 (2016). https://doi.org/10.1038/hdy.2016.45
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2016.45