Abstract
Understanding the mating patterns of populations of tree species is a key component of ex situ genetic conservation. In this study, we analysed the genetic diversity, spatial genetic structure (SGS) and mating system at the hierarchical levels of fruits and individuals as well as pollen dispersal patterns in a continuous population of Theobroma cacao in Pará State, Brazil. A total of 156 individuals in a 0.56 ha plot were mapped and genotyped for nine microsatellite loci. For the mating system analyses, 50 seeds were collected from nine seed trees by sampling five fruits per tree (10 seeds per fruit). Among the 156 individuals, 127 had unique multilocus genotypes, and the remaining were clones. The population was spatially aggregated; it demonstrated a significant SGS up to 15 m that could be attributed primarily to the presence of clones. However, the short seed dispersal distance also contributed to this pattern. Population matings occurred mainly via outcrossing, but selfing was observed in some seed trees, which indicated the presence of individual variation for self-incompatibility. The matings were also correlated, especially within (r̂p(m)=0.607) rather than among the fruits (r̂p(m)=0.099), which suggested that a small number of pollen donors fertilised each fruit. The paternity analysis suggested a high proportion of pollen migration (61.3%), although within the plot, most of the pollen dispersal encompassed short distances (28 m). The determination of these novel parameters provides the fundamental information required to establish long-term ex situ conservation strategies for this important tropical species.
Similar content being viewed by others
Introduction
Knowledge of the genetic structure, mating system and contemporary gene flow in tropical trees has increased substantially in recent years. However, most studies have focused on timber species, and little emphasis has been given to tree species bearing fruits that are valued by humans (Alves et al., 2007). Various biomes in the tropics, including the Amazon, have suffered from intense colonisation pressure and alarming rates of deforestation. Important genetic resources must be protected against these threats, and therefore, sound in situ and ex situ conservation strategies that are designed specifically for tropical conditions are urgently required.
Understanding the mating system and pollen dispersal patterns of natural populations of tree species is integral for ex situ genetic conservation because these factors determine the kinship within open-pollinated families and consequently affect the effective population size in progeny array samples. Mating systems have been shown to be dynamic across a number of tree species; the outcrossing rates and correlated matings have been shown to differ among Embothrium coccineum populations (Mathiasen et al., 2006), among individuals within populations (Platypodium elegans, Hufford and Hamrick, 2003; Magnolia stellata, Tamaki et al., 2009), among different parts of the canopy (Eucalyptus globulus, Patterson et al., 2004), among reproductive events (P. elegans, Hufford and Hamrick, 2003), and even among and within fruits within individuals (Acacia melanoxylon, Muona et al., 1991; Eucalyptus rameliana, Sampson, 1998; M. stellata, Tamaki et al., 2009). In animal-pollinated tree species, mating systems have been shown to be affected by factors such as the reproductive population density (Murawski and Hamrick, 1991) and by anthropogenic processes such as forest fragmentation (Fuchs et al., 2003; Quesada et al., 2004; Eckert et al., 2009) or logging (Obayashi et al., 2002; Lourmas et al., 2007; Lacerda et al., 2008). Because these processes reduce the density of reproductive individuals and may affect the behaviour of pollinators, the outcrossing rate and the number of mating pollen donors may decrease, which results in inbreeding and an increased relatedness within families. Consequently, the increase in inbreeding and kinship within families decreases the effective population size (Ne<4) more than that expected in populations that are characterised by random mating (Ne=4), and therefore a larger sample from a greater number of seed trees is required for ex situ conservation.
Theobroma cacao L. (cacao; Malvaceae sensu lato; Alverson et al., 1999) is an important tropical tree species that is cultivated for its valued seeds, which comprise the sole source of cocoa butter and solids for the chocolate and confectionary industries. This diploid species (2n=20) is monoecious with bisexual flowers, but it demonstrates high outcrossing rates that range from 30 to 100% (Voelcker, 1938; Benton, 1986; Efombagn et al., 2009a). Cacao is typically pollinated by midges, mainly Forcipomyia species, or by other small insects, such as ants and aphids. Two morphological and adaptive flower traits favour outcrossing in cacao: (i) the presence of a crown of staminodes (modified sterile stamens) around the stigma, which represents a physical barrier against self-pollination; and (ii) the anatomical structure of the petals, which contain a distal ligule (blade of the petal) and a proximal cowl, a shell-like modification that completely surrounds each anther (Cuatrecasas, 1964). In addition, cacao possesses a unique gameto-sporophytic self-incompatibility system (Knight and Rogers, 1955; Cope, 1976).
Incompatible mating is characterised by a failure in gametic nuclei fusion at the embryo sac that results in flower abscission (Knight and Rogers, 1955; Cope, 1962), which is considered a late-acting mechanism (Gibbs and Bianchi, 1999). The genetic control of self-incompatibility in cacao appears to be determined by a nuclear multiallelic S locus (Knight and Rogers, 1955; Cope, 1976), which is affected by other independent loci (Cope, 1958, 1962). However, the incompatibility mechanism in cacao is not strict but quantitative because it depends on the ratio of fused to non-fused ovules and individual differences in incompatibility (Cope, 1962; Warren and Kalai, 1995). The self-incompatibility mechanism in cacao can be overcome intentionally by employing a mixture of compatible and incompatible pollen with successful self-fertilisation (Glendinning, 1960) or naturally via pollination with a similar blend of pollen under field conditions. The latter conditions have been shown to result in self-pollination rates that range from 0 to 89% (Lanaud et al., 1987).
Under natural conditions, cacao seeds appear to be dispersed by animals, which mostly include small primates, rodents or birds; however, humans might be the most efficient dispersal agents. Cacao trees display a natural propensity to develop many orthotropic stems (‘suckers’) that are capable of bending in response to environmental factors. This process initiates the development of additional orthotropic shoots (Bartley, 2005). Over time, this vegetative propagation may represent a relevant dispersal mechanism, yet it still demonstrates greater restrictions in comparison to animal seed dispersal. Cacao seeds are recalcitrant, and ex situ conservation requires the maintenance of living clonal plant repositories.
The putative centre of T. cacao diversity was originally hypothesised to be located in the region between Ecuador, Colombia and Peru (Cheesman, 1944) and was later confirmed by a microsatellite marker analysis (Motamayor et al., 2002, 2008). Despite the importance of this plant, little is known about the natural genetic structure of the T. cacao population because most studies have utilised accessions that were originally collected in the wild, but maintained in ex situ germplasm repositories. The accessions have been analysed as a group according to the approximate collection location, the river basin (Sereno et al., 2006; Motamayor et al., 2008; Zhang et al., 2008). Moreover, to our knowledge, no report has estimated the clonality, intrapopulation spatial genetic structure (SGS), effective population size, mating system at the hierarchical level of fruits and individuals, and pollen flow in natural cacao populations, which are fundamental parameters that are required to establish long-term ex situ or in situ conservation strategies. Collection expeditions conducted in Amazonia have employed empirical approaches to sample budwood, seeds, or both from trees without previous definitions of the ideal number and distance between individuals for ex situ conservation sampling. The high rate of deforestation in Amazonia threatens the conservation of the untapped genetic diversity of natural cacao. In Brazil, a systematic collection project designed to obtain a representation of the genetic diversity of natural or cultivated cacao in the whole Brazilian Amazon region was conducted by the Brazilian government from 1976 to 1991 (Almeida et al., 1995), but only a small fraction of the Brazilian Amazon river basins have been sampled and maintained in ex situ collections (Sereno et al., 2006).
Therefore, we examined the genetic diversity, inbreeding, SGS and mating system at hierarchical levels of individuals and fruits within individuals and pollen dispersal patterns and distance in a continuous T. cacao population in the Brazilian Amazon. Specifically, we aimed to answer the following questions: (i) Is there a SGS in this population? (ii) Is there selfing and inbreeding in this population as observed by Efombagn et al. (2009b) for some accessions of cacao? (iii) What is the level of correlated paternity, coancestry and the effective population size within and among the fruits? (iv) What is the rate of pollen migration, and the distance and patterns of pollen dispersal in the plot? (v) What is the minimum number of seed trees necessary to collect representative seeds for conservation programmes?
Materials and methods
Study site and sampling
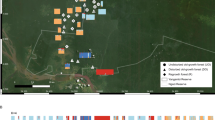
This study was conducted to evaluate a continuous cacao population in the Amazonian forest near Mocajuba, Pará State, Brazil (02°31′20.5′′ S; 49°31′62.6′′ W). The population was located in a floodplain that displayed an insular ecotype; the area was exposed to daily floods from tidewater from the Tocantins river basin caused by high tides from the ocean. This site has been used to collect cacao fruits for over 200 years, and it had been selectively logged for economic timber trees. For the study, an 80 × 70 m plot (Figure 1) was established in 2008. This plot contained 156 individuals, which were all sampled, mapped and genotyped. The cacao trees were not randomly distributed in the population, and a clear grouping was apparent in parts of the plot. No growing seedlings were found in the area, but some clonal individuals were visually identified and later confirmed by DNA analysis. Seeds from open-pollinated fruits were sampled from nine individual trees that were located close to the centre of the plot. Five fruits were sampled from each tree, and ten seeds from each fruit were used for the genetic analysis (50 seeds per seed tree for a total of 450 seeds for nine seed trees).
DNA extraction
DNA extractions and microsatellite analyses were conducted at the Molecular Biology laboratory of the ‘Comissão Executiva do Plano da Lavoura Cacaueira’ (CEPLAC), Marituba, Brazil. Young healthy leaves were collected from the trees at the site, stored in paper bags, transferred to the laboratory within two days and stored at −80 °C until extraction. DNA was extracted using a protocol adapted from Doyle and Doyle (1990), as described by Sereno et al. (2006). The DNA was quantified using a spectrophotometer (Biomate 3; Thermo Electron Co., Madison, WI, USA), and the DNA quality was evaluated by gel electrophoresis. DNA stocks were kept frozen and a working solution was prepared (5 ng μl−1).
Microsatellite analyses
Nine cacao microsatellite loci (mTcCIR3, mTcCIR12, mTcCIR25, mTcCIR35, mTcCIR37, mTcCIR75, mTcCIR100, mTcCIR223 and mTcCIR264; Lanaud et al., 1999) were used. The amplification reactions contained 15 ng of DNA, 100 μM of each dNTP, 0.2 μM of each primer and 1 U Taq polymerase (Invitrogen, São Paulo, Brazil) in 1 × PCR buffer (50 mM KCl; 10 mM Tris-HCl, pH 8.8; 0.1% Triton X-100; 1.5 mM MgCl2) in a total volume of 13 μl. The amplifications were conducted using a GeneAmp 9600 thermocycler (Applied Biosystems, Foster City, CA, USA) with the following programme: an initial cycle of 94°C for 4 min, followed by 30 cycles of 94°C for 30 s, 46°C or 51°C for 60 s and 72°C for 60 s (Lanaud et al., 1999). The amplification products were separated in denaturing sequencing gels (6% polyacrylamide; 7 M urea) that were run in 1 × TBE (Tris-Borate-EDTA) (89 mM Tris base; 89 mM boric acid; 2 mM EDTA) at 60 W for 2.5 or 3 h and visualised by silver staining (Creste et al., 2001).
Analysis of individual aggregation and clonal diversity
To verify whether the trees displayed an aggregate distribution inside the plot, we estimated the dispersal index described by Clark and Evans (1954) using SGS software (Degen et al., 2001) and the following equation: R̂CE=R̂o/R̂E, where Ro is the average distance of the closest neighbour and RE is the expected average distance of the closest neighbour. If RCE=1, RCE<1 or RCE>1, then the distribution of the individuals is considered random, aggregate or uniform, respectively. The clonal diversity was analysed according to the genotypic richness, which is estimated by R̂=(G–1)/(n−1), where G is the number of distinct genotypes and n is the number of individuals analysed (Dorken and Eckert, 2001). To determine whether repeated multilocus genotypes were clones (ramets) of the same genotype (genet), we calculated the probability of observing at least the given number of samples with the same multilocus genotypes using the following equation:

where g is the Hardy–Weinberg probability of the multilocus genotype under random mating; m is the observed number of clones (identical multilocus genotypes) and n is the total number of sampled individuals. The gene frequencies in this analysis were estimated using only unique multilocus genotypes that contained one sample of each repeated multilocus genotype (clones). If Psex<0.01, individuals with identical multilocus genotypes were ramets of the same genet. This analysis was run using GenClone 2.0 (Arnaud-Haond and Belkhir, 2007).
Genetic diversity analysis
The genetic diversities determined for adult trees (unique multilocus genotypes + one copy of each clone; n=106+21=127) and seedlings were estimated for each locus and averaged across the loci using classic genetic population parameters: the total and average number of alleles per and across loci (A) and the observed (Ho) and expected heterozygosity (He) under Hardy–Weinberg equilibrium. Inbreeding in adults was assessed by estimating the fixation index (F), and the statistical significance of the F values was tested using a 1000 Monte Carlo permutation (alleles among individuals) and a Bonferroni correction (95%, α=0.05) to avoid false positives. All parameters and permutations were estimated using Fstat (Goudet, 1995). To compare the average values of A, Ho and He between adult trees and seedlings, the 95% confidence interval (CI) of the s.e. (mean±1.96 × s.e.) for these parameters was calculated using a jackknife procedure across the loci. In the offspring, SPAGeDI version 1.3a (Hardy and Vekemans, 2002) was used to calculate the intraindividual fixation index with the reference allele frequencies that were calculated for adult trees (excluding clones; n=127). SPAGeDI version 1.3a was employed to test the significance of the F values for the seedlings using a 1000 Monte Carlo permutation of the alleles among individuals and a Bonferroni correction (95%, α=0.05).
Spatial genetic structure analysis
The intrapopulation SGS was estimated for all of the sampled individuals in the plot (n=156), for individual, unique multilocus genotypes (n=106) plus one sample of each repeated multilocus genotype (n=21) (n=127), or for clones alone (n=21+29=50). SGS was analysed by estimating the average coancestry coefficient (θxy) between all pairwise individuals in 16 distance classes of 5 m. The distance classes had at least 50 pairs of individuals within each class (in the case of clones, the minimum number of pairs was 53 and in the others analyses, this number was at least 212 pairs). Coancestry coefficients were estimated using the method of J Nason (described in Loiselle et al., 1995). To visualise SGS, θxy values were averaged over a set of distance classes and plotted against the distances (classes of 5 m, up to 80 m). To test whether there was a significant SGS, the 95% CI was calculated for each observed value, and each distance class was calculated from 1000 permutations of individuals among the locations. The CI was used to construct a coancestry graph. Coancestry coefficients and CIs were calculated using SPAGeDi version 1.3a. To compare the extension of SGS between all adult individuals (n=156), adult unique multilocus genotypes (n=127) or clonal genotypes (n=50), the Sp statistic (Vekemans and Hardy, 2004) was calculated as follows: Sp=−bk/(1−θ1), where θ1 is the average coancestry coefficient calculated between all pairwise individuals within the first distance class (0–5 m) and bk is the slope of the regression of the coancestry coefficient against the logarithm of spatial distance (up to 80 m). To test for SGS, the spatial positions of the individuals were permutated (10 000 times) to obtain the frequency distribution of bk under the null hypothesis that θ1 and ln(dxy) were uncorrelated.
Estimation of historical gene dispersal from SGS
SPAGeDi version 1.3a was used to estimate the historical gene dispersal for adults from SGS with the assumption that the observed SGS represented the equilibrium between isolation-by-distance and genetic drift (Hardy et al., 2006). The historical gene dispersal in terms of the neighbourhood size (Nb) was estimated as Nb=−(1−θ1)/bk (Vekemans and Hardy, 2004), where bk is the regression slope within the distance class σg<dij<80 m. This estimation of Nb is dependent on the value assumed for effective density, De (Hardy et al., 2006). Thus, De was estimated as De=D(Ne/N), where the effective density is the ratio of the effective population to the census population size (Vekemans and Hardy, 2004). According to previous plant studies (Hardy et al., 2006), D/10 and D/2 were adopted as a minimum and a maximum estimate of De, respectively. Fixing De, the lower and upper bounds for the 95% CI of Nb were estimated as Nb(lower)=(θ1−1)/(bk−2 s.e.b) and Nb(upper)=(θ1−1)/(bk+2 s.e.b), respectively, where s.e.b is the s.e. of bk which was calculated by jackknifing the data across each loci (Hardy et al., 2006). The 95% CI of σg was estimated as σg=√(Nb/4πDe) using the lower and upper Nb bounds (Hardy et al., 2006).
Mating system analysis
The mating system was analysed using the mixed mating model and the correlated mating model in the MLTR program, version 3.4 (Ritland, 2002). The parameters that were calculated at the population level were the multilocus outcrossing rate (tm), the single-locus outcrossing rate (ts), the selfing correlation (rs), the multilocus paternity correlation (rp(m)) and the single-locus paternity correlation (rp(s)). The difference between the single-locus and multilocus outcrossing rates (tm−ts) was used to determine whether any mating had occurred among the relatives in the population. Positive and significant differences between tm and ts were attributed to mating among relatives because ts represents the rate of mating among non-relatives, and the complement 1−ts includes apparent selfing due to mating among relatives and true selfing, and tm excludes all apparent selfing due to biparental inbreeding (Shaw et al., 1981). The parameters tm, ts, tm−ts and rp(m) were also estimated at the levels of families and fruits within families, and the multilocus paternity correlation (rp(m)) was estimated at different hierarchical levels among and within fruits. The analysis was performed at the population level using the Newton–Raphson numerical method and at the individual family and fruit levels using the Expectation–Maximization numerical method (Ritland, 2002). The 95% CI of the parameters was calculated from 1000 bootstrap permutations. In all of the analyses, the s.d. was calculated using individuals within the families as the baseline for re-sampling. The average effective number of pollen donors per seed tree and fruits was estimated as follows: N̂e(p)=1/r̂p(m) (Ritland, 1989). The average coancestry coefficient (Θ) within families was calculated from the estimator of the coefficient of relatedness within families (rxy), as derived by Ritland (1989). As the studied population was not inbred (see results), rxy was estimated according to rxy=2Θ, and the coancestry coefficient within families was estimated by dividing Ritland's estimators by two  or directly from the following expression:
or directly from the following expression:  . We also estimated the average effective population size within families (Ne) using the Cockerham (1969) estimator:
. We also estimated the average effective population size within families (Ne) using the Cockerham (1969) estimator:  , where n is the number of analysed seeds within families (n=50) or fruits (n=10) and Fo is the coefficient of inbreeding in the offspring (estimated by the fixation index). In this estimate, negative Fo values were assumed to be zero. From the effective population size within families, we estimated the number of seed trees (m) necessary for seed collection to retain the reference effective population size (Ne(reference)) of 150 (3 × 50; Lacerda et al., 2008). This estimator was calculated according to the method of Sebbenn (2003), which is based on the relationship between the effective population size goal of the conservation programme (N(e)reference), and the effective population size estimated for the average of the seed trees (Ne): m̂=N(e)reference/N̂e. This expression is based on two suppositions: (i) the seed trees are not relatives; and (ii) the seed trees do not overlap in the pollen pool. Therefore, in this case, we assumed that the seed trees were not mating with one another and were not receiving pollen from the same fathers. Thus, if related individuals are only present within families, but not among families, the Ne values of the families can be added, and a total effective population size of the progeny array can be estimated. Consequently, if the aim of the conservation programme is to retain, for example, an effective population size of 150 (N(e)reference), and the families have an effective population size represented by an average Ne, the number of seed trees required to collect seeds can be determined by dividing the reference effective population size by the average effective population size of the families: m̂=150/N̂e.
, where n is the number of analysed seeds within families (n=50) or fruits (n=10) and Fo is the coefficient of inbreeding in the offspring (estimated by the fixation index). In this estimate, negative Fo values were assumed to be zero. From the effective population size within families, we estimated the number of seed trees (m) necessary for seed collection to retain the reference effective population size (Ne(reference)) of 150 (3 × 50; Lacerda et al., 2008). This estimator was calculated according to the method of Sebbenn (2003), which is based on the relationship between the effective population size goal of the conservation programme (N(e)reference), and the effective population size estimated for the average of the seed trees (Ne): m̂=N(e)reference/N̂e. This expression is based on two suppositions: (i) the seed trees are not relatives; and (ii) the seed trees do not overlap in the pollen pool. Therefore, in this case, we assumed that the seed trees were not mating with one another and were not receiving pollen from the same fathers. Thus, if related individuals are only present within families, but not among families, the Ne values of the families can be added, and a total effective population size of the progeny array can be estimated. Consequently, if the aim of the conservation programme is to retain, for example, an effective population size of 150 (N(e)reference), and the families have an effective population size represented by an average Ne, the number of seed trees required to collect seeds can be determined by dividing the reference effective population size by the average effective population size of the families: m̂=150/N̂e.
Paternity analysis
The theoretical power to exclude the second parent (when the first is known) assuming random mating was calculated using Cervus 3.0 (Marshall et al., 1998; Kalinowski et al., 2007). The cryptic gene flow (CGF), or the probability of assigning a candidate father inside the population when the true father was outside the plot, was calculated as described by Dow and Ashley (1996): CGF=1−(1−P2°parent)N, where N is the number of candidate fathers and P2°parent is the combined non-exclusion probability of the second parent when the first (here the mother) is known. The parentage analysis was conducted based on the multilocus genotypes of the 450 seeds and all 127 unique multilocus genotypes present in the plot via a maximum-likelihood paternity assignment (Meagher, 1986) using Cervus 3.0. The most likely parental pair was determined with the Δ statistic (Marshall et al., 1998) using the reference allele frequencies that were calculated for the adult trees, as indicated by Meagher and Thompson (1987). To determine the putative father of the seeds, all 127 genets were tested as putative paternal parent candidates. The significance of Δ was determined using paternity tests that were simulated by the software (critical Δ) using a confidence level of 80%, a genotyping error ratio of 0.01 and 50 000 repetitions. The calculation of critical Δ values was based on the assumption that 90% of the sampled candidates were located within the plot. We also evaluated self-fertilisation. If a father candidate had a Δ value higher than the critical Δ value calculated by the simulations, it was considered the true parent. If the same individual was found to be the paternal parent, this seedling was considered selfed. Thus, the selfing rate (s) was estimated as the number of selfed seeds divided by the total number of analysed seeds. The pollen immigration rate (m) was calculated as the ratio of seeds that had no parents inside the plot relative to the total number of sampled seeds. Because all of the sampled individuals had known spatial positions, the effective pollen dispersal distance was calculated from the position of the seed tree relative to its putative father. Because of the presence of clones in the population, the distance between the seed tree and each identical clone that was determined to be the father of a seed was averaged. To investigate whether mating success was a function of the distance between trees, we compared the frequency distribution of the realised pollination with the frequency distribution of the distances among all trees using the Kolmogorov–Smirnov test (Sokal and Rohlf, 1995).
Results
Clonal diversity
The dispersal index (RCE=0.83) estimation indicated that the distribution of the individuals within the plot was aggregated (Figure 1). The observed average distance of the closest neighbour was 2.41 m. The analysis of clonal diversity indicated that among the 156 trees analysed, 29 individuals (R̂=0.81) were clones (ramets, defined as vegetatively propagated copies of a tree) from 21 genets (unique multilocus genotypes with more than one individual within the plot) among a total of 50 clones (n=29+21) in the plot, whereas the remaining 106 trees were unique multilocus genotypes. The probability of observing at least the given number of samples with the same multilocus genotypes under random mating was low (Psex<0.001; or 1:1000), which allowed the determination of identical multilocus genotypes as ramets of the genets. The number of ramets per ortet ranged from one to four.
Genetic diversity and inbreeding
From the nine loci analysed, only one (mTcCIR100) was monomorphic, and this locus was excluded from the subsequent genetic analyses. The population presented a low-polymorphism level at these eight loci (Table 1). The total number of alleles detected across the eight loci was 26 either for the adult trees (n=127 unique multilocus genotypes) or for the seeds analysed. No unique alleles were detected among the adults and seeds. The average number of alleles per locus (Â=3.25) was the same for adults and seeds, which suggested that pollen migration did not occur in the plot or that the gene pool of the population was represented in the plot. The average observed heterozygosity was significantly higher in the adults (Ĥo=0.496) than in the seeds (Ĥo=0.391), according to the 95% CI calculated by jackknifing across the loci. The same trend was observed for the expected heterozygosity, and a significantly higher value was determined for the adult trees (Ĥe=0.477) in comparison to the seeds (Ĥe=0.408). The fixation index was not significantly different from zero for the adult trees. Conversely, the fixation index for five of the eight loci (mTcCIR3, mTcCIR25, mTcCIR35, mTcCIR37 and mTcCIR75) and the overall average demonstrated an increase in homozygosity in the seeds relative to the adults, although this increase was significantly different from zero for only two loci (mTcCIR37 and mTcCIR75) and the overall mean, which suggested that inbreeding had occurred among the offspring. In contrast, for two other loci (mTcCIR12 and mTcCIR64), the fixation indices were positive in the adults and negative in the seeds; for one locus (mTcCIR12), the estimated value was significantly <0, which suggested the presence of selection for heterozygosity. Assuming that microsatellite loci are theoretically selection neutral, it is possible that this locus is linked to a locus that is undergoing selection, which could explain the inconsistent results. When locus mTcCIR12 was excluded from the analysis, the fixation index increased to 0.121 (P<0.05), which was close to the inbreeding level expected for mating among half-sibs.
Intrapopulation spatial genetic structure
The SGS was analysed for three distinct subsamples: (i) all of the genotypes (n=156); (ii) the genets alone (unique multilocus genotypes, n=127) or (iii) the clones alone (n=21+29=50). A strong significant intrapopulation SGS was observed for the population (Figure 2), especially when either all genotypes or the clone subsamples were analysed. In all the analyses, the coancestry coefficient decreased with increasing distance between the individuals, and it was significant for the first three distance classes (0–5, 5–10 and 10–15 m). This finding indicated that closer individuals were more genetically similar than was expected based on the random distribution. When the analysis included clones alone, the coancestry in the first class (0–5 m) was 0.220, and this value was higher than that obtained for genets alone (θ̂xy=0.0465). This result indicated that the occurrence of clones had a strong impact on the SGS. The distance among clones ranged from 0.4 to 12.1 m and demonstrated an average of 3.8±2.1 m (±95% s.e.). The expected coancestry between two clones that originated from an outcrossing was 0.5, and our estimation of coancestry between pairwise clones ranged from 0.04 to 1.0 and demonstrated an average of 0.44±0.25 (±95% s.e.). The regression slope bk for the pairwise coancestry coefficient over the logarithm of spatial distance (0–80 m) was significantly negative for all of the analyses (Table 2), which indicated the presence of isolation by distance. The intensity of SGS, as measured by the Sp statistic for clones alone (n=50; Sp=0.0320), was higher than that for the genets (n=127; Sp=0.0179) or for all individuals (n=156, Sp=0.0209).
Intrapopulation spatial genetic structure in a Theobroma cacao population. (a) All individuals (n=156); (b) adults without clones (n=132); (c) clonal population (n=50). The solid line represents the average θxy value. The dashed lines represent the 95% (two-tailed) confidence interval of the average θxy distribution, which was calculated based on 1000 permutations of the spatial distance among pairs of adults.
Historical gene dispersal
The estimate for the historical gene dispersal distance (Table 2) based on the regression slope (bk; 0–80 m) that were calculated for the total sample (n=156) ranged from 19 m (assuming De=D/2) to 36 m (assuming De=D/10). The estimate for clones alone was 18 m (assuming De=D/2). For the unique multilocus genotypes, the estimates for σg did not converge for both assumed effective densities, whereas for the clones alone, the estimate for σg did not converge under the assumption of De=D/10 (Table 2). The estimates for neighbourhood size (Nb) ranged from 14 (clones) to 48 (adults; Table 2).
Mating system
The estimated correlation of selfing was high (r̂s=0.508), which suggested that there was a high variation among the individual outcrossing rates (Table 3). Consistent with this observation, the estimate for the multilocus outcrossing rate (tm) varied among individual seed trees from 0.42 to 1.00 (Table 4) and demonstrated a population average of 0.964 (Table 3). For the population average and for four seed trees, the estimated outcrossing rate was significantly (P<0.05) <1 (Tables 3 and 4). The average population single-locus outcrossing rate was also significantly <1 (ts=0.935, P<0.05, Table 3). We also observed variation among the fruits within seed trees for the multilocus outcrossing rate (ranging from 0.26 to 1.0; Table 4). The difference tm−ts was positive and significantly different from zero for five seed trees (ranging from 0.06 to 0.09), whereas it was estimated to be 0.030 for the population average, which suggested that some mating among relatives occurred (Tables 3 and 4). The multilocus paternity correlation (rp(m)) was also highly variable among the seed trees (ranging from 0.03 to 0.79) and among the fruits within the seed trees (ranging from 0.01 to 0.96), which indicated that some of the seeds from the same seed tree were full-sibs (Table 4). The population multilocus paternity correlation (r̂p(m)aw=0.191; Table 3) was significantly less than the single-locus paternity correlation (r̂p(s)aw=0.387), which suggested that some of the pollen donors were relatives. Based on the multilocus paternity correlation, which was estimated at the individual and population levels (Tables 3 and 4), we estimated that between 1.27 and 33.33 pollen donors (average of 5.24) mated with the sampled seed trees. The coancestry coefficient within families (Θ) varied among the seed trees from 0.134 to 0.324 (average of 0.152). The effective population size (Ne) within the families ranged from 1.53 to 3.54 among the seed trees (average of 2.93) (Tables 3 and 4). Therefore, for the seed samples to represent an effective population size of 150, it would be necessary to collect seeds from at least 51 seed trees.
Correlated mating among and within fruits
To investigate the variation in self-incompatibility among individual trees and the kinship among and within fruits within individuals, the present seed sample was collected at hierarchical levels among and within the fruits. We estimated the outcrossing rate, the rate of mating among relatives, the paternity correlation, the effective number of pollen donors, the coancestry coefficient, the effective population size within families and the number of seed trees necessary to collect seeds within and among fruits (Tables 3 and 4). The multilocus paternity correlation was significantly higher within fruits (r̂p(m)=0.607) than among fruits (r̂p(m)=0.099), which indicated that the probability of collecting two full-sib seeds within a fruit was higher than the probability that the seeds were collected from different fruits from the same seed tree. The effective numbers of pollen donors were 1.65 and 10.1 within fruits and among fruits within seed trees, respectively (Table 3). The average coancestry coefficients were 0.200 and 0.142 within and among fruits, respectively. Consequently, the average effective population size was 2.32 within fruits and 3.11 among fruits, which revealed the requirement for seeds collection from at least 65 seed trees (Table 3), if seeds were to be collected from a single fruit per seed tree, and from 48 seed trees, if seeds were to be collected from many fruits of each seed tree to reach the reference effective population size of 150.
Gene flow
The low polymorphism level detected for the analysed loci resulted in a relatively low-exclusion power for the second parent (P(Ex2)=0.91463). Therefore, the estimate for cryptic gene flow was high (99%), which indicated a high probability of detecting a paternal seed parent within the plot when the true paternal parent was in fact outside the plot. From 450 sampled seeds, only 174 (38.7%) had a father that was within the plot (Table 4). These 174 seeds were apparently fathered by 58 (43.9%) of the potential male parents in the plot, and each male fathered from 1 to 12 seeds. Among these 174 seeds, 11 (2.4%) were produced by selfing or by relatives with indistinguishable multilocus genotypes (clones), which suggested the presence of a selfing rate of 2.4% (11/450; Table 5).
Based on the 174 seeds with a pollen donor that was determined to be inside the plot, the pollen dispersal distance ranged from 0.2 to 67 m (average and median of 28 m and 28 m, respectively; Table 5, Figure 3). The average pollen dispersal distance ranged from 14.3 to 46.5 m among the seed trees (Table 5). These distances were shorter than the average distance among all 156 trees relative to the 9 seed trees analysed (38±20 m; ranging from 0.2 to 98 m), which suggested a pattern of short pollen dispersal within the plot. The curve for the frequency of effective pollen dispersal was significantly different from that for the frequency of distance among all 156 trees relative to the 9 seed trees (D=0.184; P=0.00006; Figure 3). This result demonstrated that the distance among the trees did not explain the patterns of pollen dispersal within the plot.
Discussion
Genetic diversity
Cacao populations in the upper Amazon region have been suggested to possess a greater genetic diversity in comparison to those in the lower Amazon (Sereno et al., 2006; Motamayor et al., 2008). The genetic diversity observed for the present plot comprising a continuous stand of cacao trees under natural conditions in the lower Amazon region supports this hypothesis. The expected heterozygosity observed in the adults (He=0.477) was less than that previously detected in other lower Amazon populations (for example, He=0.553; Sereno et al., 2006), whereas both populations demonstrated lower values than that detected in populations in the upper Amazon region, such as the Huallaga (He=0.610) and Ucayali (He=0.740) valleys in the Peru or the coastal valley of Ecuador (He=0.561, Zhang et al., 2008). In comparison, other studies investigated more diverse cacao germplasm from the upper Amazon region or from the hybrid Trinitario genetic group and detected greater genetic diversity (He=0.620, Efombagn et al., 2009a) than that determined herein. One possible explanation for the lower expected heterozygosity observed herein could be related to the origin of the population, which was typically the lower Amazon Amelonado (Motamayor et al., 2008). However, the results obtained with less precise method of allele detection using silver-stained polyacrylamide gels cannot be discounted, as the other studies have utilised fluorescence capillary electrophoresis (Zhang et al., 2008; Efombagn et al., 2009b). Furthermore, the low level of genetic diversity in the present plot could also be explained by a small number of founder individuals followed by genetic drift.
Intrapopulation SGS
The plot population presented an aggregated spatial distribution that was associated with genetic aggregation. A strong SGS was detected, especially for the clones (Figure 2), which suggested that the seeds and propagules were dispersed a short distance. As expected using a model of isolation by distance, the coancestry coefficient progressively decreased from the first distance classes to the final ones, which indicated that closer individuals (up to 15 m) were likely related individuals or ramets from the same genet. The clonality analysis showed that the clones were spaced from 0.4 to 12 m apart within the distance of the detected SGS. However, when the clones were excluded from the analysis, SGS was also detected, but the coancestry was reduced to half. This result indicated that related individuals that had probably originated from a short seed dispersal distance were present within the plot. Although a short seed dispersal distance contributed to the SGS, vegetative propagation at short distances was likely the main cause of the observed SGS, as evidenced by the detection of the strongest SGS in the analysis based on clones alone.
The vegetative propagation of T. cacao might benefit individual fitness by avoiding haphazard losses due to fallen trees that comprise the upper stories or other environmental disturbances. A fallen cacao trunk is able to produce orthotropic shoots, which will eventually root and potentially develop into an independent individual following the decay of old tissues. This process of regeneration was clearly detected during the fieldwork and confirmed by genotyping. This clonal propagation could also lead to increases in tree density and in the dispersal surface. A cacao trunk may reach up to 20 m in height under natural conditions, and a fallen trunk may allow the development of a series of clonal plants from the same genotype, that reach dispersal distances up to the height of the original tree.
Mating system
The present mating system analysis indicated that T. cacao might produce seeds mainly via outcrossing (Tables 3 and 4). However, selfing was observed in some of the seed trees and fruits within the seed trees. The estimated population selfing rate (ŝ=1−t̂m=1–0.964=0.036) was within the reported ranges of selfing for self-incompatible cacao accessions (3–8%), whereas selfing rates ranging from 25 to 43% were detected for self-compatible genotypes (Yamada and Guries, 1998). Mixtures of outcrossing and selfing cacao seeds have also been documented by Efombagn et al. (2009b), who used a paternity analysis and microsatellite loci to detect an average of 38% selfing; among seed trees, selfing ranged from 0 to 75%.
The presence of different levels of incompatibility in self-incompatible individuals has been well documented in cacao (Glendinning, 1960; Cope, 1962; Lanaud et al., 1987; Warren and Kalai, 1995). These variations in the selfing rate of self-incompatible cacao may be partially attributed to a mechanism that overcomes incompatibility via natural pollination with mixtures of compatible (‘pollen mentor’) and self-incompatible pollen (Glendinning, 1960) or a genotypic effect (Aneja et al., 1994).
The major consequence of the intrapopulation spatial genetic structure in natural cacao populations is the possible occurrence of matings among relatives. Our estimates for tm−ts provided significant evidence for mating among relatives in five seed trees, many fruits within seed trees, and for the population average (Tables 3 and 4). Interestingly, these five seed trees (Table 4) were the individuals with the highest outcrossing rates (>0.98), and they appeared to be strongly self-incompatible. These results suggested that selfing did not take place in these trees, but mating among relatives might have occurred.
Biparental inbreeding in a generation is equivalent to the coancestry of the parental population. We estimated inbreeding in the seeds and observed an inbreeding level that might have been produced by mating among first cousins or half-sibs (Table 1).
Correlated mating
This is the first study to correlate mating in a T. cacao population. Our results demonstrated high levels of variation in the correlated matings among individuals and fruits within seed trees (Tables 3 and 4). A high rate of correlated mating has also been observed for the self-incompatible congener tree species Theobroma grandiflorum under natural conditions (Alves et al., 2003). The correlated paternity within and among fruits depends primarily on the reproductive structures and mating system of the species (Muona et al., 1991). For animal-pollinated tree species, insect vectors may deposit several pollen grains obtained from the most recently visited parent tree, which could result in full-sib seeds within a fruit (Muona et al., 1991). Correlated mating may also occur when mating occurs primarily between near neighbours, and both processes may occur in this cacao population. However, the present results suggested that the observed high-paternity correlation within fruits was more likely due to the transfer of multiple pollen grains via one pollinator visit rather than a short distance pollen dispersal, which would imply equal probabilities of multiple paternity among and within fruits.
The frequencies of correlated matings were higher within than among fruits (Table 3), which suggested that a restricted number of effective pollen donors were the fathers of the seeds within fruits (Nep=1.65), although a large number of pollen donors mated with each seed tree (Nep=10.1). Therefore, there was a greater probability of obtaining two full-sib seeds from a single fruit (60.7%) than from different fruits in a single sampling. Thus, most of the fruits appeared to contain full-sibs. A higher paternity correlation within rather than among fruits has been detected in other plant species (Muona et al., 1991; Sampson, 1998; Quesada et al., 2001; Tamaki et al., 2009).
Pollen flow and dispersal
A high proportion (average of 61%) of the pollen that fertilised the nine seed trees was obtained from trees located outside the plot. However, according to our estimation of the cryptic pollen flow, the pollen migration rates might have been higher than the estimated values, and the average pollen dispersal distance was likely underestimated. In contrast, pollen from inside the plot was dispersed at a high frequency across short distances (70% up to 40 m, an average of 28 m). Considering the seeds with putative fathers that were identified within the plot, the maximum pollen dispersal distance was estimated to be 67 m, which was likely an underestimation due to the increased pollen migration. This value was consistent with the indirect estimation of the flight distance of cacao pollinator midges (Forcipomyia), which cover distances of up to 60 m (Van Der Knaap, 1955). The prevalence of cross-pollination at distances of up to 40 m has also been estimated by Benton (1986).
Pollen was not randomly dispersed within the plot, and the distance between trees in the plot did not explain the observed patterns of pollen dispersal. It has been established that cross-pollination in cacao occurs preferentially among neighbouring trees (Harland and Flechville, 1927; Posnette, 1950). According to Yamada and Guries (1998), the pollinator Forcipomyia typically travels between adjacent trees, and pollination occurs within a range from 3 to 12 m; however, pollination can occur at a distance of up to 40 m, but this process occurs at a low frequency unless the individual densities in the area are low. In fact, the results suggested that there were two patterns of pollen dispersal: a short distance and a long distance. The short pollen dispersal distance may be explained by the behaviour of the pollen dispersers (Forcipomyia midges), which prefer to visit flowers among close neighbouring trees. The longer pollen dispersal distance may be explained by temporal variation in the flower phenology. For example, if only distant trees are flowering, the pollinators must travel long distances to locate resources. However, to better understand long-distance pollen dispersal in T. cacao, larger plots must be assessed in future studies. For populations with the same density of reproductive trees (>150 ha−1), square plots of ∼4 ha that contain ∼600 potential pollen donors will likely be adequate; plots of this size will increase the chances of finding a larger number of pollen donors inside the plot and consequently enable researchers to construct more accurate pollen-dispersal curves.
Implications for breeding and conservation sampling
The effective number of pollen donors (Nep) varied widely among the families and among the fruits (89% of the fruits were pollinated by <6 pollen donors; Table 4). These low numbers of pollen donors that mated with the seed trees increased the coancestry coefficient within families and consequently decreased the effective population size because of an increased frequency of alleles that were identical by descent. The coancestry coefficient within families and among and within fruits was higher than that expected for true half-sib families (Θ=0.125, Table 4). Within fruits, Θ was closer to the value expected for full-sib families (Θ=0.200; Table 3), whereas among fruits, it was closer to the values expected for half-sib families (Θ=0.142; Table 3). Therefore, the effective population size within fruits (Ne=2.32; Table 3) was lower than that among fruits (Ne=3.11; Table 3). Consequently, the seed collection strategies for ex situ conservation and breeding must consider the mixing of seeds from a larger number of fruits from each seed tree to maximise the Ne of the seed lots. Consistent with our results, using this strategy, the coancestry within families will decrease to values that are closer to that expected for true half-sib families (Θ=0.125), and the Ne of each family will be closer to the maximum value that a family can achieve (Ne=4).
For tree breeding and conservation genetic strategies, it is important to establish the spatial distance and number of seed trees for the seed collection. Information concerning clonality, SGS, the mating system and the pollen dispersal distance may facilitate these recommendations. If seed trees are selected within the spatial scale of the SGS, there is a possibility that they are relatives. If seeds from related seed trees are collected, the frequency of identical alleles by descent increases among families, and therefore, the Ne of the total sample (mixture of seeds from different seed trees) decreases. Thus, because we detected SGS at distances up to 15 m, the selected seed trees in the studied population must be located at distances >15 m between each other. However, although this sampling strategy may prevent the selection of relative seed trees, it does not preclude the possibility that seeds from distinct seed trees will be relatives due to mating among these trees (full-sibs between families) or because different seed trees will receive pollen from the same pollen pool.
Based on the analysis of the pollen dispersal distance, we observed that ∼31% of the pollen was dispersed inside the plot at distances up to 67 m. Thus, to prevent the collection of seeds from seed trees that mate with themselves and/or overlap in the pollen pool, seed trees located at distances >67 m must be selected for seed collections. The present results did not permit the proposition of a precise recommendation because of an underestimation of the pollen dispersal distance. However, the ideal distance is likely >67 m. This important point must be investigated in future studies of plots that are larger than the one assessed herein.
Finally, the present results demonstrated that in collections from many fruits from each seed tree, the reference effective population size of 150 could be achieved when seeds were collected from ∼50 seed trees. However, it is important to note that this is an underestimation because it is based on the assumption that there is no relatedness between families and that the seed trees used to collect seeds were not exposed to overlapping pollen pools. These assumptions were likely to be violated in the present population because many of the sampled seed trees received overlapping pollen pools and mated among themselves. Despite this finding, the realised estimate can be used as a minimum number of seed trees from which to collect seeds for breeding and conservations purpose.
References
Almeida CMVC, Machado PFR, Barriga JP, Silva FCO (1995). Coleta de cacau (Theobroma cacao L.) da Amazônia brasileira: uma abordagem histórica e analítica.. Ministério de Agricultura e Reforma Agrária: Comissão Executiva do Plano da Lavoura Cacaueira, Boletim Técnico: Belém, Pará, Brazil. pp. 92.
Alverson WS, whitlock BA, Nyffeler R, Bayer C, Baum DA (1999). Phylogeny of the core Malvales: evidence from ndhF sequence data. Am J Bot 89: 1474–1486.
Alves RM, Sebbenn AM, Artero AS, Clement C, Figueira A (2003). Mating system in natural population of Theobroma grandiflorum (Willd ex Spreng) Shumm. Genet Mol Biol 26: 373–379.
Alves RM, Sebbenn AM, Artero AS, Clement C, Figueira A (2007). High levels of genetic divergence and inbreeding in populations of cupuassu (Theobroma grandiflorum). Tree Genet Gen 3: 289–298.
Aneja M, Gianfagna T, Ng E, Badilla I (1994). Carbon dioxide treatment partially overcomes self-incompatibility in a cacao genotype. HortScience 29: 15–17.
Arnaud-Haond S, Belkhir K (2007). GENCLONE: a computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Mol Ecol Notes 7: 15–17.
Bartley BGD (2005). The Genetic Diversity of Cacao and its Utilization. CABI Publishing: Oxfordshire, UK, 341 p.
Benton FP (1986). Distância Alcançada por Insetos Polinizadores na Transferência de Pólen do Cacaueiro. CEPLAC, Informe de Pesquisas: Ilhéus. pp. 57–58.
Clark DA, Evans FC (1954). Distance to nearest neighbour as a measure of spatial relationship in populations. Ecology 35: 445–456.
Cheesman EE (1944). Notes on the nomenclature, classification and possible relationships of cocoa populations. Trop Agric 21: 144–159.
Cockerham CC (1969). Variance of gene frequencies. Evolution 23: 72–84.
Cope FW (1958). Incompatibility in Theobroma cacao. Nature 181: 279.
Cope WF (1962). The mechanism of pollen incompatibility in the Theobroma cacao L. Heredity 17: 157–182.
Cope FW (1976). Cacao, Theobroma cacao. In: Simmonds NW (ed) Evolution of Crop Plants.. Longman: London. pp 285–289.
Creste S, Tulmann-Neto A, Figueira A (2001). Detection of single sequence repeat polymorphisms in denaturing polyacrylamide sequencing gels by silver staining. Pl Mol Biol Report 19: 299–306.
Cuatrecasas J (1964). Cacao and its allies: a taxonomic revision of the genus Theobroma. Contrib US Natl Herbarium 35: 379–614.
Degen B, Petit R, Kremer A (2001). SGS—Spatial Genetic Software: a computer program for analysis of spatial genetic and phenotypic structure of individuals and populations. J Heredity 92: 447–448.
Dorken ME, Eckert CG (2001). Severely reduced sexual reproduction in northern populations of a clonal plant, Decodon verticillatus (Lythraceae). J Ecology 89: 339–350.
Dow BD, Ashley MV (1996). Microsatellite analysis of seed dispersal and parentage of sampling in bur oak, Quercus macrocarpa. Mol Ecol 5: 615–627.
Doyle JJ, Doyle JL (1990). Isolation of plant DNA from fresh tissue. Focus 12: 13–15.
Efombagn IB, Motamayor JC, Sounigo O, Eskes AB, Nyassé S, Cilas C et al. (2009a). Genetic diversity and structure of farm and Genbank accessions of cacao (Theobroma cacao L.) in Cameroon revealed by microsatellite markers. Tree Genet Genomes 4: 821–831.
Efombagn IB, Sounigo O, Eskes AB, Motamayor JC, Manzanares-Dauleux MJ, Schnell R et al. (2009b). Parentage analysis and outcrossing patterns in cacao (Theobroma cacao L.) farms in Cameroon. Heredity 103: 46–53.
Eckert CG, Kalisz S, Geber MA, Sargent R, Elle E, Cheptou P-O et al. (2009). Plant mating system in a changing world. Trends Ecol Evol 25: 35–43.
Fuchs EJ, Lobo JA, Quesada M (2003). Effects of forest fragmentation and flowering phenology on the reproductive success and mating patterns of the tropical dry forest tree Pachira quinata. Conserv Biol 17: 149–157.
Gibbs PE, Bianchi MB (1999). Does late-acting self-incompatibility (LSI) show family clustering? Two more species of Bignoniaceae with LSI: Dolichandra cynanchoides and Tabebuia nodosa. Ann Bot 84: 449–457.
Glendinning DR (1960). Selfing of self-incompatible cacao. Nature 187: 170.
Goudet J (1995). Fstat. (Version 2.9.3.2.): a computer program to calculate F-statistics. J Heredity 86: 485–486.
Hardy O, Vekemans X (2002). SPAGeDI: a versatile computer program to analyze spatial genetic structure at the individual or population levels. Mol Ecol Notes 2: 618–620.
Hardy OJ, Maggia L, Bandou E, Breyne P, Caron J, Chevallier MH et al. (2006). Fine-scale genetic structure and gene dispersal inferences in 10 Neotropical tree species. Mol Ecol 15: 559–571.
Harland SC, Flechville GE (1927). Natural crossing and the genetics of axil spot in cacao. Genética 9: 279–288.
Hufford KM, Hamrick JL (2003). Viability selection at three early life stages of the tropical tree, Platypodium elegans (Fabaceae, Papilionoideae). Evolution 57: 518–526.
Kalinowski ST, Taper ML, Marshall TC (2007). Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16: 1099–1106.
Knight R, Rogers H (1955). Incompatibility in Theobroma cacao. Heredity 9: 67–69.
Lacerda EBL, Kanashiro M, Sebbenn AM (2008). Long-pollen movement and deviation of random mating in a low-density continuous population of Hymenaea courbaril in the Brazilian Amazon. Biotropica 40: 462–470.
Lanaud C, Risterucci AM, Pieretti I, Falque M, Bouet A, Lagoda PJL (1999). Isolation and characterization of microsattelites in Theobroma cacao L. Mol Ecol 8: 2141–2143.
Lanaud C, Sounigo O, Amefia YK, Paulin D, Lachenaud P, Clément D (1987). New data on the mechanisms of incompatibility in cocoa and its consequences on breeding. Café Cacao Thé 31: 278–282.
Loiselle BA, Sork VL, Nason J, Graham C (1995). Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am J Bot 82: 1420–1425.
Lourmas M, Kjellberg F, Dessard H, Joly HI, Chevallier M-H (2007). Reduced density due to logging and its consequences on mating system and pollen flow in the African mahogany Entandrophragma cylindricum. Heredity 99: 151–160.
Marshall TC, Slate J, Kruuk LEB, Pemberton JM (1998). Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol 7: 639–655.
Mathiasen P, Rovere AE, Premoli AC (2006). Genetic structure and early of inbreeding in fragmented temperate forest of a self-incompatible tree, Embothrium coccineum. Conserv Biol 21: 232–240.
Meagher TR (1986). Analysis of paternity within a natural population of Chamaelirium luteum.1. Identification of most-likely male parents. Am Nat 128: 199–215.
Meagher TR, Thompson E (1987). Analysis of parentage for naturally established seedlings of Chamaelirim luteum (Liliaceae). Ecology 68: 803–812.
Motamayor JC, Risterucci AM, Lopez PA, Ortiz CF, Moreno A, Lanaud C (2002). Cacao domestication I: the origin of the cacao cultivated by the Mayas. Heredity 89: 380–386.
Motamayor JC, Lachenaud P, Mota JWS, Loor R, Kuhn DN, Brown S et al. (2008). Geographic and genetic population differentiation of the Amazonian chocolate tree (Theobroma cacao L). PLoS ONE 3: e3311.
Muona O, Moran GF, Bell JC (1991). Hierarchical patterns of correlated mating in Acacia melanoxylon. Genetics 127: 619–626.
Murawski DA, Hamrick JL (1991). The effect of the density of flowering individuals on the mating systems of nine tropical tree species. Heredity 67: 167–174.
Obayashi K, Kalasa J, Armesto JJ, Collins SL (2002). The ecological concept of disturbance and its expression at various hierarquical levels. Inter J Plant Sci 163: 151–158.
Patterson B, Vaillancourt RE, Pilbeam DJ, Potts BM (2004). Factors affecting variation in outcrossing rate in Eucalyptus globules. Aust J Bot 54: 773–780.
Posnette AF (1950). The pollination of cacao in the Gold Coast. J Hort Scien 25: 155–163.
Quesada M, Fuchs E, Lobo J (2001). Pollen load size, reproductive success and progeny kinship of natural pollinated flowers of the tropical dry forest tree, Pachira quinta. Am J Bot 88: 2113–2118.
Quesada M, Stoner KR, Lobo JA, Herrerías-Diego Y, Guevara CQ, Munguía-Rosas A et al. (2004). Effects of forest fragmentation on pollinator activity and consequences for plant reproductive success and mating patterns in bat-pollinated Bombacaceous trees. Biotropica 36: 131–138.
Ritland K (1989). Correlated matings in the partial selfer Mimulus guttatus. Evolution 43: 848–859.
Ritland K (2002). Estimation of gene frequency and heterozygosity from pooled samples. Mol Ecol Notes 78: 370–372.
Sampson JF (1998). Miltiple parternity in Eucalyptus rameliana (Myrytaceae). Heredity 81: 349–355.
Sebbenn AM (2003). Tamanho amostral para conservação ex situ de espécies arbóreas com sistema misto de reprodução. Rev Inst Flor 15: 109–124.
Sereno ML, Albuquerque PSB, Vencovsky R, Figueira A (2006). Genetic diversity and natural population structure of cacao (Theobroma cacao L.) from the Brazilian Amazon evaluated for microsatellite markers. Cons Genet 7: 13–24.
Shaw DV, Kahler AL, Allard RW (1981). A multilocus estimator of mating system parameters in plant populations. Proc Natl Acad Sci USA 78: 1298–1302.
Sokal RR, Rohlf FJ (1995). Biometry: Principles and Practices of Statistics in Biological Research. Third Edition. W.H. Freeman and Company: New York.
Tamaki I, Setsuko S, Tomaru N (2009). Estimation of outcrossing rates at hierachical levels of fruits, individuals, populations and species in Magnolia stellata. Heredity 102: 381–388.
van Der Knaap WP (1955). Observations on the pollination of cacao flowers. Hort Congs 2: 1287–1293.
Vekemans X, Hardy OJ (2004). New insights from fine-scale spatial genetic structure analysis in plant populations. Mol Ecol 13: 921–935.
Voelcker OJ (1938). The incidence of cross-pollination in cacao. Seventh Annual Report on Cacao Research (Trinidad) pp. 9–14.
Warren J, Kalai SM (1995). Isozyme markers for self-compatibility and yield in Theobroma cacao (cacao). Heredity 74: 354–356.
Yamada MM, Guries RP (1998). Mating system analysis in cacao (Theobroma cacao L.). Agrotrópica 10: 165–176.
Zhang D, Boccara M, Motilal L, Butler DR, Umaharan P, Mischke S et al. (2008). Microsatellite variation and population structure in the ‘Refractario’ cacao of Ecuador. Cons Genet 9: 327–337.
Acknowledgements
We thank Dr Kermit Ritland for assistance with the MLTR program and technician Admil de Souza Moura for excellent technical assistance. We also acknowledge the financial support from FAPESPA and CNPq, which provided a grant and fellowships, respectively. We are grateful to the three reviewers for thoroughly reading, correcting and providing suggestions for our manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Silva, C., Albuquerque, P., Ervedosa, F. et al. Understanding the genetic diversity, spatial genetic structure and mating system at the hierarchical levels of fruits and individuals of a continuous Theobroma cacao population from the Brazilian Amazon. Heredity 106, 973–985 (2011). https://doi.org/10.1038/hdy.2010.145
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2010.145
Keywords
This article is cited by
-
Human influence on the distribution of cacao: insights from remote sensing and biogeography
Biodiversity and Conservation (2024)
-
Phenology, Seed Germination, and Genetics Explains the Reproductive Strategies of Diospyros lasiocalyx (Mart.) B. Wall
Tropical Plant Biology (2020)
-
Short distance pollen dispersal and low genetic diversity in a subcanopy tropical rainforest tree, Fontainea picrosperma (Euphorbiaceae)
Heredity (2019)
-
Demographic history and spatial genetic structure in a remnant population of the subtropical tree Anadenanthera colubrina var. cebil (Griseb.) Altschul (Fabaceae)
Annals of Forest Science (2019)
-
Stepping stones or stone dead? Fecundity, pollen dispersal and mating patterns of roadside Qualea grandiflora Mart. trees
Conservation Genetics (2019)