Abstract
Pseudovivipary is an environmentally induced flowering abnormality in which vegetative shoots replace seminiferous (sexual) inflorescences. Pseudovivipary is usually retained in transplantation experiments, indicating that the trait is not solely induced by the growing environment. Pseudovivipary is the defining characteristic of Festuca vivipara, and arguably the only feature separating this species from its closest seminiferous relative, Festuca ovina. We performed phylogenetic and population genetic analysis on sympatric F. ovina and F. vivipara samples to establish whether pseudovivipary is an adaptive trait that accurately defines the separation of genetically distinct Festuca species. Chloroplast and nuclear marker-based analyses revealed that variation at a geographical level can exceed that between F. vivipara and F. ovina. We deduced that F. vivipara is a recent species that frequently arises independently within F. ovina populations and has not accumulated significant genetic differentiation from its progenitor. We inferred local gene flow between the species. We identified one amplified fragment length polymorphism marker that may be linked to a pseudovivipary-related region of the genome, and several other markers provide evidence of regional local adaptation in Festuca populations. We conclude that F. vivipara can only be appropriately recognized as a morphologically and ecologically distinct species; it lacks genetic differentiation from its relatives. This is the first report of a ‘failure in normal flowering development’ that repeatedly appears to be adaptive, such that the trait responsible for species recognition constantly reappears on a local basis.
Similar content being viewed by others
Introduction
Environmental conditions, especially temperature, rainfall and photoperiod, vary with time and location. Plants can change their phenotype in tune with variations in environmental conditions. In some cases, these adaptive changes are genetically controlled such that the progeny of individuals from different environments retain their different characteristics when grown together. If this differential adaptation to the environment occurs within a species, races with distinct ecological preferences emerge (Jump et al., 2006). These races can in turn evolve into new species.
In many temperate grasses, marginal floral inductive conditions (mainly photoperiod and temperature) induce proliferation of inflorescences in which flowering is aborted and leafy shoots are produced instead of sexual reproductive structures (Arber, 1934; Beetle, 1980) (Figure 1). This phenotype, known as pseudovivipary, generally prevails in wet areas, mainly in alpine and arctic regions, where the leafy propagules can survive and establish successfully (Wycherley, 1953a, 1954; Lee and Harmer, 1980; Heide, 1994; Elmqvist and Cox, 1996). Pseudovivipary is thought to be a genetically assimilated form of asexual reproduction that arises from habitually seed-producing (seminiferous) species (Heide, 1988).
Comparison of F. ovina and F. vivipara spikes and spikelets. (a, b) F. ovina spike and spikelet (sp); the spike is made up of spikelets of three florets each. The spikelet meristem terminates in an empty lemma (el) (arrow). (c, d) In F. vivipara spikelets, florets are replaced by shoots. Scale bar=5 mm. g, glume; l-like, leaf-like lemma.
The distribution of constantly pseudoviviparous forms often overlaps with that of closely related seminiferous taxa, with the asexual forms typically occupying environments that do not favour the production of viable seed. Examples of such sympatric seminiferous–pseudoviviparous pairings include Festuca vivipara (L.) Sm. with Festuca. ovina L. sensu stricto (hereafter termed F. ovina), and Poa bulbosa L. sensu stricto with Poa bulbosa var. vivipara Koel. (Wycherley, 1953a; Beetle, 1980; Heide, 1989). The stability of the pseudoviviparous trait after transplantation to a common growing environment (Youngner, 1960; Moore and Doggett, 1976; Beetle, 1980; Heide, 1988, 1989, 1994) suggests a genetic rather than a plastic (environmentally induced) maintenance of the phenotype. Grass forms exhibiting phenotypically fixed pseudovivipary are generally taxonomically recognized at infraspecific ranks, although there is little consensus between taxa and researchers (Wycherley, 1953b; Youngner, 1960; Beetle, 1980). F. vivipara is an exception to this rule, as it is a species whose defining characteristic is pseudovivipary (Beetle, 1980), although the taxon has been recognized at several infraspecific ranks (Wycherley, 1953b; Beetle, 1980; Frederiksen, 1981; Wilkinson and Stace, 1991). Researchers have postulated that F. vivipara arose from F. ovina through mutation, hybridization and/or polyploidy (Arber, 1934; Flovik, 1938; Wycherley, 1953b; Beetle, 1980).
Pseudovivipary can be experimentally induced in a wide range of seminiferous grasses by growing them under suboptimal floral inductive conditions; moreover, in facultative pseudoviviparous forms, sexual reproduction can be partially restored under optimal flowering conditions (Moore and Doggett, 1976; Heide, 1989). In addition, pseudoviviparous plants produce fertile flowers in areas in which pseudoviviparous and seminiferous populations overlap occasionally (Wycherley, 1953a, 1954; Lee and Harmer, 1980). This phenomenon, present in a number of species of Poa and Festuca, indicates that sexuality is not totally suppressed in all pseudoviviparous individuals (Wycherley, 1953a, 1954; Beetle, 1980; Lee and Harmer, 1980; Heide, 1988). These observations led Beetle (1980) to suggest that pseudoviviparous forms arose in response to prolonged exposure to conditions favourable for their development. Thereafter, these forms spread to occupy their current range. According to this hypothesis, pseudoviviparous members of the same species share a monophyletic origin. The alternative, polyphyletic theory proposes that pseudovivipary is constantly evolving among seminiferous populations that are found in conditions that are not conducive for normal flowering (Wilkinson and Stace, 1991). Determining the actual origin of pseudovivipary is central to establishing a common practice for the taxonomic treatment of these taxa, and for understanding the ecological and evolutionary role of the phenomenon (Wilkinson and Stace, 1991).
Thus far, studies of pseudovivipary have focused on describing the phenotype and the conditions that induce it, and on exploring its ecological significance (Wycherley, 1954; Beetle, 1980; Lee and Harmer, 1980; Heide, 1994). Although information about the cause(s) of pseudovivipary has been widely reported, little is known about how it is controlled and maintained at a molecular or genetic level (Wycherley, 1953b; Youngner, 1960; Beetle, 1980; Lee and Harmer, 1980; Heide, 1989). Determining the phylogeographic origin of pseudoviviparous grasses is a first natural step towards answering this question. To do this, we performed a phylogeographic study of F. vivipara and sympatric F. ovina from the United Kingdom using amplified fragment length polymorphism (AFLP) and sequence variation of chloroplast DNA loci (cpDNA).
We integrated population genetic and phylogenetic analysis to explain the geographic distribution of genealogical lineages among F. ovina and F. vivipara samples, and to investigate the genetic polymorphism linked to pseudovivipary. Similar approaches, relying on the correlation between gene clines and selective environmental transition, have been used in a range of situations (Beaumont and Nichols, 1996; Beaumont and Balding, 2004; Jump et al., 2006; Bonin et al., 2007; Caballero et al., 2008; Herrera and Bazaga, 2008). Our data are more consistent with the hypothesis that UK F. vivipara repeatedly arises de novo from local seminiferous populations of F. ovina.
Materials and methods
Plant material
Paired samples of F. vivipara (L.) Sm. with F. ovina L. sensu stricto plants (growing within 5 m of each other) were collected from over 50 localities around three mountains in the United Kingdom: Ben Lawers (Mid Perthshire, Scotland), Snowdon (Caernarvonshire, North Wales) and Craig Cerrig Gleisiad (Breconshire, South Wales) (Supplementary Figure 1). At locations at which either F. ovina or F. vivipara was absent, samples of the species present were collected. In total, more than 120 samples were collected.
Samples used to assemble cpDNA sequence data comprised of 48 individuals from Snowdon, 23 from Ben Lawers and 9 from Craig Cerrig Gleisiad. The AFLP analysis included 26 samples from Snowdon, 16 from Ben Lawers and 14 from Craig Cerrig Gleisiad. All plant samples were confirmed to be either F. vivipara or F. ovina by morphological and ploidy level analysis (Wilkinson and Stace, 1991).
DNA extraction
DNA was extracted from 0.1 g of fresh leaves using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). The samples used for both cpDNA and AFLP analyses were chosen to represent all three collection sites, and wherever possible to include paired neighbouring samples of F. vivipara and F. ovina. DNA quality and quantity were assessed using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies LLC, Wilmington, DE, USA), and the concentration adjusted to 5–10 ng μl−1 for cpDNA sequencing and to 10–15 ng μl−1 for AFLP analysis.
Chloroplast DNA sequence analysis
Initially, three non-coding cpDNA regions, trnCF-rpoB, trnS(GCU)-psbD and trnT2-rps4 (Saltonstall, 2001), were amplified and assessed for sequence variation among the following eight samples: four F. vivipara and four F. ovina from Snowdon and Ben Lawers using primers published by Saltonstall (2001). These cpDNA regions have been previously shown to be polymorphic in Festuca pratensis (Fjellheim et al., 2006). For PCR, 25 μl reaction mixes comprising 12.5 μl Biomix (Bioline, London, UK), 25 pmol of each primer and 2.5 μl of DNA template were assembled. The reaction mix was then subjected to 1 min at 94 °C, followed by 35 cycles of 30 s at 94 °C, 40 s at 56 °C and 1.5 min at 72 °C, culminating with 10 min at 72 °C. PCR amplicons were sequenced directly, twice in both directions, using the amplification primers after cleaning with a NucleoFast 96 PCR Cleanup kit (Macherey-Nalgene, Düren, Germany). Cleaned PCR products were sent to Macrogen Inc. (Korea) for cycle sequencing and product fractionation using dye terminator chemistry with an ABI3730XL unit (Sanger et al., 1977; Macrogen Inc., Seoul, Korea). Sequences were aligned using ClustalW2 (Larkin et al., 2007) and the alignments edited using MacClade 4.07 software package (Maddison and Maddison, 2002; Sinauer Associates, Sunderland, MA, USA).
Following the initial screen, the two variable loci (trnCF-rpoB and trnT2-rps4) were used to obtain sequence in both directions for the remaining samples. For the larger screen, cpDNA sequence data were also obtained from GenBank for Agrostis stolonifera, F. pratensis haplotype A, B and C (from Georgia, Bulgaria and the United Kingdom, respectively), Norwegian Festuca gigantea, Festuca arundinacea and Lolium perenne; these were used as reference samples for the phylogenetic analysis (Fjellheim et al., 2006; Saski et al., 2007). A. stolonifera was used as the out-group sample.
Parsimony analysis to construct a phylogeny was performed using Phylogenetic Analysis Using Parsimony*4.0b10 (Swofford, 1998). Heuristic parsimony search was conducted using 100 random addition sequence replicates and tree bisection reconnection branch swapping, retaining no more than 10 trees per replicate. Bootstrap analysis was carried out using Phylogenetic Analysis Using Parsimony implementing the same heuristic search strategy and character type settings as in the phylogeny construction and 100 bootstrap replicates. The structure of the tree results from Phylogenetic Analysis Using Parsimony was further explored in MacClade. The level and distribution of genetic variation in F. ovina and F. vivipara populations observed from cpDNA sequence data was further explored in AFLP analysis.
AFLP analysis
Fluorescent-AFLP analysis was performed using the AFLP Core Reagents and AFLP Primer Starter kits (Invitrogen, Paisley, UK). Selective amplicons generated using 6-carboxyfluorescein (FAM)-and 6-carboxy-2′,4,4′,5′,7,7′-hexachlorofluorescein (HEX)-labelled primers were fractionated by capillary electrophoresis on an ABI PRISM 3100 capillary electrophoresis unit. Initially, a total of six primer pairs were screened: (1) M-CAA:E-ACG, (2) M-CTC:E-ACG, (3) M-CTC:E-GAC*, (4) M-CAT:E-GAC*, (5) M-CAT:E-GCA, (6) M-CAA: E-GCAFAM*. Three of these (asterisked) that produced consistent and scorable profiles were selected for further analysis. Electropherogram traces were analysed using GeneMarker Version 1.71 (Softgenetics LCC, State College, PA, USA).
AFLP profiles were scored for the presence or absence of amplicon peaks between 80 and 500 bp in size using GeneMarker software (Softgenetics LCC). Peaks of similar size and intensity were assumed to be homologous.
AFLP marker diversity among F. ovina/F. vivipara populations
To estimate genetic diversity and population differentiation, analysis of molecular variance (AMOVA) was carried out on the binary data set derived from GeneMarker using GenAlex software, version 6.2 (Peakall and Smouse, 2006; The Australian National University, Canberra, Australia). We used the binary genetic distance option, which results in the phiPT estimator, an analogue of FST. Six populations defined by mountain and species, pooled into three regions defined by geographic origin were compared in this analysis. Principal coordinate analysis was used to graphically represent the genetic distance matrices generated from AMOVA.
Identification of candidate loci for selection
Outlier loci (those deemed potentially under selection) were identified using DFDIST software (www.rubric.rdg.ac.uk/~mab/stuff/) (Beaumont and Nichols, 1996). The sub-populations compared with DFDIST were as follows: (1) Three populations defined by mountain (F. ovina and F. vivipara pooled together on each mountain); (2) For F. ovina, three populations defined by mountain; (3) For F. vivipara, three populations defined by mountain; and (4) For each mountain, two populations of F. ovina and F. vivipara. A total of 50 000 realizations were performed using the default settings of the DFDIST program with a smoothing proportion of 0.04, target average FST equal to the trimmed mean FST (from Ddatacal software, distributed with DFDIST; http://www.rubic.rdg.ac.uk/~mab/stuff/), a θ-value of 0.06 and maximum allowable frequency of 0.99 pooled across samples. The Benjamini and Hochberg false discovery rate (FDR) correction method was used to correct for the occurrence of false positives in loci identified as under selection (Benjamini and Hochberg, 1995). The P-values used in the FDR analysis were derived from the three P-values (P1, P2 and P3) produced by the pv program (distributed with DFDIST; http://www.rubic.rdg.ac.uk/~mab/stuff/). The reason DFDIST produces three P-values is because of the presence of ties in the data. The empirical probability of observing simulated FST values as small as or smaller than the data point is given by P1, and the probability of observing values greater than or equal to the data point is given by P2. A third P-value, P3, is also given, which is defined as follows: if P1⩾0.5, then the greater of P1 and P2; if P1<0.5, the lesser of P1 and P2. However, in this study, we used Px, computed as Px=P1−0.5(P1+P2−1), which was then converted into a two-tailed P-value by computing 1–2 × abs(Px−0.5) (Beaumont and Nichols, 1996; Beaumont and Balding, 2004). We used Px because it is based on the midpoint of the ties, and has a relatively uniform simulated distribution, in line with the assumption of methods that compute the FDR. Loci with significant P-values at a FDR threshold of 50% were identified using the Benjamini and Hochberg method.
Results
Genetic diversity and differentiation among F. ovina/F. vivipara populations
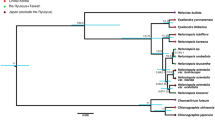
Chloroplast sequence analysis: The cpDNA sequence alignment produced a data matrix of 1055 characters. In the strict consensus tree from parsimony analysis, the samples were separated into four major clades: group 1, 2 and 3 containing all F. ovina and F. vivipara samples, and group 4 with all other Festuca spp. and L. perenne samples from GenBank (Figure 2). The Festuca ovina-vivipara samples under study separated into group 3, comprising F. vivipara samples from Craig Cerrig Gleisiad and Snowdon; group 2, comprising F. vivipara samples from Ben Lawers and one F. ovina sample from Snowdon; and group 1 comprising the remaining samples. The F. ovina/F. vivipara samples were found to comprise 10 chloroplast haplotypes, separated among them by between 1 and 10 unambiguous changes, and collectively separated by more than 10 changes from samples in group 4 (Figure 2b). Representative sequences for each haplotype were submitted to the GenBank genetic sequence database (trnCF-rpoB and trnT2-rps4, respectively): S18p—accession numbers HM807585 and HM807595; S10s—accession numbers HM807586 and HM807596; CC7s—accession numbers HM807587 and HM807597; S37p—accession numbers HM807588 and HM807598; S26s—accession numbers HM807589 and HM807599; S18s—accession numbers HM807590 and HM807600; S37p—accession numbers HM807591 and HM807601; BL4p—accession numbers HM807592 and HM807602; S10s—accession numbers HM807593 and HM807603; BL2p—accession numbers HM807594 and HM807604.
Results of parsimony analysis of Festuca cpDNA sequence data. (a) Strict consensus tree from parsimony analysis of cpDNA sequence data. The samples are separated into four groups: groups 1–4. Groups 1, 2 and 3 had modest bootstrap support. Scale bar represents one change. (b) Consensus tree from (Figure 1a) condensed in MacClade. Groups 1 and 2 are separated from group 3 by 10 unambiguous changes, whereas F. ovina and F. vivipara samples in groups 1 and 2 are separated by one or two base changes. Horizontal bars represent the number of unambiguous changes on each branch. BL, Ben Lawers; CC, Craig Cerrig Gleisiad; p, F. vivipara; s, F. ovina; S, Snowdon.
It was clear from the cpDNA data that the F. ovina/F. vivipara complex is separate from the other Festuca spp. included in the study. The chloroplast haplotypes observed here could not be used to demarcate clades defined by species (F. vivipara or F. ovina). The strongly supported clusters recovered from the analysis were moderately concordant with geographic origin, but not with species identity. Therefore, we further explored the genetic diversity and differentiation among the F. ovina/F. vivipara populations using AFLP analysis.
AFLP analysis: We generated 105 repeatable AFLP bands (loci) of consistent intensity for the 56 samples used in the study. Of the 105 loci, 65 were found to be polymorphic between samples. The AFLP data were used to explore the level of genetic diversity and its distribution among the Festuca populations.
Genetic differentiation among Festuca populations: AMOVA with six populations, defined by sample origin and species, and three regions, defined by the mountains, showed that 5% of the AFLP variation among samples was due to differences between the mountains, 3% due to variation among the populations and the remaining 92% was due to variation within populations (Table 1).
In almost all the pairwise comparisons between the six populations, variation was highly significant (P<0.01) between populations from different locations. Within locations, significant variation was only observed between F. ovina and F. vivipara among samples from Craig Cerrig Gleisiad (Table 2).
The results from AMOVA were supported by a plot of the first two principal coordinates extracted from the principal coordinate analysis (Figure 3a). Here, F. ovina and F. vivipara samples from the same mountain did not form distinct clusters and, whereas Ben Lawers and Snowdon samples were separate from each other, the Craig Cerrig Gleisiad cluster overlapped with the former two. The plot is reminiscent of the phylogenetic tree based on cpDNA variation, as the samples separated into location-based groups separated by statistically significant genetic differentiation.
Analysis of AFLP variation among Festuca populations. (a) A plot of Festuca samples from Snowdon (S), Ben Lawers (BL), and Craig Cerrig Gleisiad (CC) produced from principal coordinate analysis. p, pseudoviviparous; s, seminiferous. The percentages of total variation accounted for by the first two principal coordinates (PCO 1 and PCO 2) are shown in brackets. (b) Comparison of the distribution of per-locus FST of F. ovina and F. vivipara samples from Craig Cerrig Gleisiad (n=14). Estimated values of FST from 56 AFLP loci are plotted as a function of heterozygosity. In (b–d), solid lines denote 0.995th, 0.5th and 0.005th quantiles of the conditional distribution obtained from DFDIST simulation and outlying loci are highlighted in black. (c) Comparison of the distribution of per-locus FST of F. vivipara samples from Snowdon, Ben Lawers and Craig Cerrig Gleisiad (n=32). Estimated values of FST from 64 AFLP loci are plotted as a function of heterozygosity. (d) Comparison of the distribution of per-locus FST of F. ovina and F. vivipara samples from Snowdon, Ben Lawers and Craig Cerrig Gleisiad (n=55). Estimated values of FST from 65 AFLP loci are plotted as a function of heterozygosity.
Identification of outlier loci: Further examination of the AFLP data using DFDIST sought to determine whether, given the relatively low differentiation between populations of F. ovina and F. vivipara within the same mountain, there was evidence of any highly differentiated loci. In the within-mountain comparisons carried out here, an outlier locus was only detected among Craig Cerrig Gleisiad samples (Figure 3b). In addition, this locus (50) was only an outlier in this comparison, not detected in other comparisons (Figures 3b–d), and was well within the 50% FDR threshold (P=0.006).
With samples subdivided into three populations per species (defined by mountain of origin), outlier loci were only observed from the F. vivipara comparisons: loci 63, 2, 24 and 3 fell within the 50% threshold in FDR analysis with P-values of 0.004, 0.005, 0.006 and 0.016 (Figure 3c). One of the loci detected here (locus 2) was the same as that detected in an analysis in which the species were pooled into one population for each mountain (Figures 3c and d). A similar analysis with all F. ovina and F. vivipara samples pooled together revealed no outliers at the 50% FDR threshold, although locus 2 was borderline (P=0.008). Thus, we find evidence of adaptive divergence among mountains at some loci in F. vivipara, but not in F. ovina. Furthermore, evidence of adaptive divergence at these loci disappears when the two species are pooled within each mountain.
Discussion
Several authors have suggested that, in most cases, pseudovivipary is merely a floral or inflorescence reversion that arises spontaneously in several grass species in montane-artic conditions and that has unclear ecological or taxonomic significance (Beetle, 1980; Lee and Harmer, 1980). Only in F. vivipara has the trait led to the definition of a new species. We investigated the genealogical and population genetic relationships between sympatric stands of F. vivipara and seminiferous F. ovina to gain insights into the genetic control of pseudovivipary and the role it has in speciation.
F. vivipara and F. ovina are not genetically distinct
The phylogeny of pseudoviviparous and seminiferous Festuca plants from three locations across the United Kingdom, compiled on the basis of two intergenic chloroplast loci (trnT2-rps4 and trnCF-rpoB), revealed only a low level of variation between samples, with just 10 distinct haplotypes observed in total. Nevertheless, there was sufficient structuring between locations to allow a loose classification based on geographic origin, but not to consistently distinguish F. ovina from F. vivipara. Although more regions of the chloroplast could be usefully deployed to further resolve the sequence-based phylogeny, it is likely that most additional loci that could be used for this purpose would be less informative than those already selected. For this reason, we elected to use AFLP data to provide finer resolution of genetic relationships between samples.
In common with the cpDNA sequence data, the AFLP data revealed separation of samples into groups on the basis of geographical origin, supported by AMOVA, principal coordinate analysis and DFDIST analysis. Similar levels of differentiation could not be detected between the two Festuca species. The AFLP data failed to clearly separate samples according to species identity for all but one mountain studied here. This is in line with the cpDNA sequence data. The results point to significant levels of genetic exchange between the two species, making it impossible to distinguish between them, except at Craig Cerrig Gleisiad, where one locus was highly differentiated between the two species. Although this locus could be a false positive, it provides evidence for a genetic basis for the differentiation between F. vivipara and F. ovina at one site. The fact that this species-specific polymorphism does not hold in the two other sites clearly indicates that the genetic variant is not itself directly causal of pseudovivipary. Nevertheless, it does not rule out the possibility that this marker may be linked to such a locus. Under this scenario, it can be speculated that directional selection around this locus leads to variation in the mode of reproduction among F. ovina plants, giving rise to de novo formation of F. vivipara. It would be interesting to observe whether this inference holds, or more loci can be identified, with a larger sample size. If the inference holds, characterization of this and similar loci may ultimately lead to the identification of the genetic basis of pseudovivipary.
Adaptive differentiation and speciation
The species-by-species analysis demonstrated that it is possible to distinguish F. vivipara populations from different locations on the basis of both cpDNA and AFLP data. Analysis of AFLP data shows that genetic exchange is greater among F. ovina than among F. vivipara plants, an observation clearly in agreement with the two species’ modes of reproduction. Interestingly, the locus identified as distinguishing between F. vivipara and F. ovina (locus 50) is not recovered as an outlier in intermountain comparisons. This suggests that there were genetic differences between populations of F. vivipara progenitors (F. ovina) from the three different locations, as shown by loci 2, 3, 24 and 63, but some of this has been lost over time. Because of limited gene flow between populations, only allowed by the occasional flowering of F. vivipara (Wycherley, 1954; Wilkinson and Stace, 1991), most of the genetic differences have been retained by local F. vivipara populations. This deduction provides further support for the local evolution of F. vivipara from F. ovina. Therefore, it would appear that, on encountering conditions unfavourable for normal flowering, F. ovina is replaced by F. vivipara, not from other areas but from within its population. There exist in F. ovina stands small amounts of genetic differentiation linked to propensity to pseudoviviparous development, for example, around locus 50 in this analysis. Montane-arctic conditions would clearly confer a selective advantage to individuals possessing this propensity, leading to their proportionate increase in these environments. For these newly established populations to continue to adapt to montane-arctic conditions, which are unfavourable to flowering, there is a presumed requirement for gene flow from other similarly adapted populations (Jump et al., 2006). This does not happen, however, as our DFDIST analysis of F. vivipara populations revealed; there is limited genetic exchange between F. vivipara populations from geographically separate locations. This limit on genetic exchange makes long-term genetic fixation of the pseudoviviparous trait impossible and could explain why habitually pseudoviviparous grasses partially revert to normal flowering under conducive flowering conditions (Heide, 1989). The mountain-by-mountain AFLP data analysis thus suggests that F. vivipara is constantly arising from F. ovina locally.
F. vivipara is a morphologically and ecologically distinct species
The low levels of differentiation between seminiferous and pseudoviviparous Festuca plants are consistent with a polyphyletic origin for F. vivipara. A polyphyletic origin does not necessarily preclude species recognition, however, as many hybrids accorded recognition at specific rank are nevertheless known to have polyphyletic origins, for example, Helianthus deserticola (Gross et al., 2003). F. vivipara is common in areas of high rainfall and low sunlight, which include rocky crevices, and rocky scree slopes (Wycherley, 1953a; Wilkinson and Stace, 1991). These conditions are thought to be conducive for the survival and establishment of F. vivipara's non-dormant, leafy propagules while making impossible regular sexual reproduction in seminiferous grass species (Wycherley, 1953a; Lee and Harmer, 1980). In this study, we did not uncover evidence of any consistent genetic differentiation that separates F. ovina from F. vivipara. This does not necessarily mean that F. vivipara is not a species, as there are no set levels of genetic differentiation required to support species-level taxonomic delineation (Lowe et al., 2004). Even though these two Festuca taxa are not genetically different and lack complete reproductive isolation from each other, they can be considered to be different species separated by ecological and morphological differentiation (Lowe et al., 2004). Although the cpDNA and AFLP data analyses suggest that F. vivipara is not defined by major evolutionary genetic processes, ecological processes that make impossible sexual reproduction have clearly led to its development from F. ovina. In addition, F. vivipara can be defined as a morphological species; when F. vivipara and F. ovina are grown under common environmental conditions, they remain morphologically distinct.
In summary, we demonstrate a lack of clear genetic differentiation between F. vivipara and F. ovina. This is coupled with a shared genetic structuring indicating repeated local emergence of pseudovivipary from seminiferous F. ovina. Furthermore, the limited geographic scale over which genetic differentiation becomes apparent between F. ovina and F. vivipara suggests a high propensity for transition from seminiferous to pseudoviviparous states. For this reason, we argue that the link between this high propensity to proliferate and spikelet meristem reversion warrants further investigation.
References
Arber A (1934). The Graminaeae: A Study of Cereal, Bamboo and Grasses. Cambridge University Press: Cambridge.
Beaumont MA, Balding DJ (2004). Identifying adaptive genetic divergence among populations from genome scans. Mol Ecol 13: 969–980.
Beaumont MA, Nichols RA (1996). Evaluating loci for use in the genetic analysis of population structure. 263: 1619–1626.
Beetle AA (1980). Vivipary, proliferation, and phyllody in grasses. J Range Manage 33: 256–261.
Benjamini Y, Hochberg Y (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodol) 57: 289–300.
Bonin A, Ehrich D, Manel S (2007). Statistical analysis of amplified fragment length polymorphism data: a toolbox for molecular ecologists and evolutionists. Mol Ecol 16: 3737–3758.
Caballero A, Quesada H, Rolan-Alvarez E (2008). Impact of amplified fragment length polymorphism size homoplasy on the estimation of population genetic diversity and the detection of selective loci. Genetics 179: 539–554.
Elmqvist T, Cox PA (1996). The evolution of vivipary in flowering plants. Oikos 77: 3–9.
Fjellheim S, Rognli OA, Fosnes K, Brochmann C (2006). Phylogeographical history of the widespread meadow fescue (Festuca pratensis Huds.) inferred from chloroplast DNA sequences. J Biogeogr 33: 1470–1478.
Flovik K (1938). Cytological studies of arctic grasses. Hereditas 24: 265–376.
Frederiksen S (1981). Festuca vivipara (Poaceae) in the North Atlantic area. Nord J Bot 1: 277–292.
Gross BL, Schwarzbach AE, Rieseberg LH (2003). Origin(s) of the diploid hybrid species Helianthus deserticola (Asteraceae). Am J Bot 90: 1708–1719.
Heide OM (1988). Environmental modification of flowering and viviparous proliferation in Festuca vivipara and Festuca ovina. Oikos 51: 171–178.
Heide OM (1989). Environmental control of flowering and viviparous proliferation in seminiferous and viviparous arctic populations of two Poa species. Arctic Alpine Res 21: 305–315.
Heide OM (1994). Control of flowering and reproduction in temperate grasses. New Phytol 128: 347–362.
Herrera CM, Bazaga P (2008). Population-genomic approach reveals adaptive floral divergence in discrete populations of a hawk moth-pollinated violet. Mol Ecol 17: 5378–5390.
Jump AS, Hunt JM, Martinez-Izquierdo JA, Penuelas J (2006). Natural selection and climate change: temperature-linked spatial and temporal trends in gene frequency in Fagus sylvatica. Mol Ecol 15: 3469–3480.
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H et al (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948.
Lee JA, Harmer R (1980). Vivipary, a reproductive strategy in response to environmental stress. Oikos 35: 254–265.
Lowe A, Harris S, Ashton P (2004). Ecological Genetics: Design, Analysis, and Application. Blackwell Publishing Company: Oxford.
Maddison DR, Maddison WP (2002). Macclade: Analysis of Phylogeny and Character Evolution. Sinauer Associates Inc.: Sunderland, MA.
Moore DM, Doggett MC (1976). Pseudovivipary in Fuegian and Falkland Islands grasses. Br Antarct Surv Bull 43: 103–110.
Peakall R, Smouse PE (2006). GENALEX 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6: 288–295.
Saltonstall K (2001). A set of primers for amplification of noncoding regions of chloroplast DNA in the grasses. Mol Ecol Notes 1: 76–78.
Sanger F, Nicklen S, Coulson AR (1977). DNA sequencing with chain-terminating inhibitors. Proc Nat Acad Sci USA 74: 5463–5467.
Saski C, Lee S-B, Fjellheim S, Guda C, Jansen R, Luo H et al (2007). Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera, and comparative analyses with other grass genomes. Theor Appl Genet 115: 571–590.
Swofford DL (1998). PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods). Sinauer Associates: Sunderland, MA.
Wilkinson MJ, Stace CA (1991). A new taxonomic treatment of the Festuca ovina L aggregate (Poaceae) in the British Isles. Bot J Linn Soc 106: 347–397.
Wycherley PR (1953a). The distribution of the viviparous grasses in Great Britain. J Ecol 41: 275–288.
Wycherley PR (1953b). Proliferation of spikelets in British grasses: I The taxonomy of viviparous races. Watsonia 3: 41–56.
Wycherley PR (1954). Vegetative proliferation of floral spikelets in British grasses. Ann Bot 18: 119–127.
Youngner VB (1960). Environmental control of initiation of the inflorescence, reproductive structures, and proliferations in Poa bulbosa. Am J Bot 47: 753–757.
Acknowledgements
We thank the following people from the School of Biological Sciences, University of Reading, for their assistance during this study: Ronald Rutherford and Stephen Jury (sample collection and identification); Mike Shaw, Julie Hawkins and Alastair Culham (genetic analysis). We are grateful to the University of Reading for financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Heredity website
Supplementary information
Rights and permissions
About this article
Cite this article
Chiurugwi, T., Beaumont, M., Wilkinson, M. et al. Adaptive divergence and speciation among sexual and pseudoviviparous populations of Festuca. Heredity 106, 854–861 (2011). https://doi.org/10.1038/hdy.2010.128
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2010.128
Keywords
This article is cited by
-
Systemic fungal endophytes and ploidy level in Festuca vivipara populations in North European Islands
Plant Systematics and Evolution (2014)
-
Fungal endophyte mediated occurrence of seminiferous and pseudoviviparous panicles in Festuca rubra
Fungal Diversity (2014)
-
Overexpression of a Transcription Factor OsMADS15 Modifies Plant Architecture and Flowering Time in Rice (Oryza sativa L.)
Plant Molecular Biology Reporter (2012)