Abstract
The complex transformation of a tadpole to a frog during amphibian development is under the control of thyroid hormone (T3). T3 is known to regulate gene transcription through its nuclear receptors. We have previously isolated many genes which are up-regulated by T3 in the intestine of Xenopus laevis tadpoles. We have now cloned a full-length cDNA for one such gene (IU12). Sequence analysis shows that the IU12 cDNA encodes a plasma membrane protein with 12 transmembrane domains and homologous to a mammalian gene associated with cell activation and organ development. Similarly, we have found that IU12 is activated during intestinal remodeling when both cell death and proliferation take place. Furthermore, IU12 is an early T3-response gene and its expression in the intestine during T3-induced metamorphosis mimics that during normal development. These results argue for a role of IU12 in the signal transduction pathways leading to intestinal metamorphosis.
Similar content being viewed by others
Introduction
Organogenesis is a critical step during postembryonic vertebrate development. It involves complex but coordinated proliferation and differentiation of various types of cells that specify a given organ. Compared to our understanding of early embryogenesis, relatively little is known about postembryonic organ development. This is in part due to the lack of proper model systems.
Amphibian metamorphosis offers several unique features for studying organogenesis and tissue remodeling1, 2, 3. This process systematically transforms various organs of a tadpole and concurrently generates adult frog organs that are highly similar to those in mammals4, 5, 6. Furthermore, extensive investigations since the beginning of the century have accumulated an enormous amount of morphological and biochemical information about the metamorphic transformations of various organs or tissues4, 5, 6, 7, 8. Finally, this process is absolutely dependent upon thyroid hormone (T3, 3, 5, 3′-triiodothyronine)4, 5, 9, 10. This has made it possible to easily manipulate metamorphosis by simply controlling the availability of T3.
The cloning of nuclear thyroid hormone receptors (TRs) and the demonstration that TRs are transcription factors11, 12, 13, 14, 15 have greatly stimulated the molecular studies on amphibian metamorphosis. Thus, T3 is believed to bind to TRs, which in turn regulate the transcription of target genes to effect metamorphosis. Therefore, it becomes critical to identify the genes regulated by T3 and study their regulation and function during metamorphosis.
Using various approaches including subtractive differential screening, different laboratories have isolated many T3-regulated genes during amphibian metamorphosis2, 16, 17. Included among them are genes which appear to be involved in the remodeling of the Xenopus tadpole intestine. Here we report the detailed characterization of one such intestinal gene. Full-length cDNA cloning reveals that this gene encodes a transmembrane protein homologous to a mammalian protein associated with cell activation. Similarly, we demonstrate here that this gene is activated by T3 during the proliferation of adult cells in the intestine, implicating a role in the development and maintenance of the adult intestine.
Materials and Methods
Isolation and characterization of Xenopus IU12 full-length cDNA clone
The small IU12 (Intestinal Up-regulated fragment #12) cDNA fragment isolated from the subtractive hybridization library18 was used as a probe to screen for full length cDNA clones from a library made from intestinal mRNA19. A clone with a 2.3 kb insert was isolated, sequenced by using Sequenase v.2.0 kit (US Biochemical), and determined to be the Xenopus homology of a human gene associated with cell activation. The Xenopus IU12 sequence reported in this paper is has been deposited into GenBank with the accession number AFO 19906.
Treatment of X. laevis tadpoles and RNA isolation
About 20-40 tadpoles at stages 52-5420 were treated in 1 liter of dechlorinated tap water with 5nM (T3). The solution was changed daily. Whenever a treatment lasted > 2 d, the tadpoles were fed continuously. The protein synthesis inhibitors cyclohexamide and anisomycin (CHX) were added at 20 and 25 μg/ml, respectively, for 13 h beginning 1 h before the addition of T3. This treatment inhibits protein synthesis in tadpole tissues by 99%21. RNA was isolated as described22. The entire tadpole intestinal tract except the stomach region was isolated for analysis.
Northern blot analysis
Total RNA was electrophoresed on a 1% agarose formaldehyde gel and transferred onto a Gene Screen membrane (NEN) after partial hydrolysis with NaOH23, 24. Hybridization was done overnight at 42 °C in 50% formamide, 5X SSPE, 0.2% SDS, 10% Dextran sulfate, 5X Denhardt's solution, and 100 μg/ml denatured salmon sperm DNA, and the filters were washed three times for 5-10 min each at room temperature in 2X SSC and 0. 2% SDS. Stringent washes were then carried out twice for 25 min each in 0.25X SSC and 0.2% SDS at 65 °C.
To control for RNA loading and quality, the Northern blots were stained with methylene blue before hybridization25. In addition, they were hybridized with the cDNA for the ribosomal protein L8 (rpL8), whose expression is constant during development and independent of T326.
PCR Southern blot analysis of tissues specific expression
Southern blots were prepared from PCR amplified cDNAs made of mRNAs from the limb, intestine, tail, and brain of stage 52-54 tadpoles treated with or without T3 for 1 d18, 27, 28, 29. The blots were hybridized with IU12 cDNA or rpL8 as a control.
Sequence analysis
Sequence homology searches were done by using the GCG program. For prediction of protein localization sites, the PSORT program (http://psort.nibb.ac.jp/form.html) was employed.
Results
In an effort to study the molecular events underlying the thyroid hormone regulation of amphibian metamorphosis, we have previously performed a PCR-based subtractive differential screen to isolate genes which are regulated by T3 in metamorphosing intestine18. This resulted in the isolation of over 20 small PCR cDNA fragments derived from genes whose mRNA levels were up-regulated in the intestine within 24 h of T3-treatment of premetamorphic tadpoles. In this paper, we focus on the cloning, sequence analysis, and developmental expression of one such T3-response gene, i.e., the gene IU1218.
Cloning and sequence analysis of Xenopus IU12 cDNA
To determine the identity of the T3-response gene IU12, the small PCR fragment was used to screen a Lambda cDNA library made of intestinal mRNA. A cDNA clone with a 2.3 kb insert was thus isolated. Conceptual translation of the cDNA revealed the existence of an open reading frame encoding a polypeptide of 507 aa in length (Fig 1). The existence of upstream in-frame stop colons indicates that the first methionine is the true amino terminus of the polypeptide. The cDNA has a 270 bp 5′-untranslated region and a 460 bp 3′-untranslated region. Interestingly, it lacks a consensus polyadenlylation signals, suggesting that the cDNA clone was derived from an alternatively polyadenlylated mRNA or due to the internal priming of the full-length mRNA by the dTn primer during the cDNA library construction. In agreement with this, the full-length mRNA is about 5 kb (see below), much longer than the cDNA insert. Thus, the full length mRNA most likely contains a very long 3′-untranslated region, like many other T3-response genes isolated from the metamorphosing Xenopus laevis16, 19, 28, 30, 31.
Sequence analysis by using the PSORT (prediction of protein calization sites) program revealed the existence of 12 transmembrane domains (Fig 1A), all of which are highly hydrophobic (Fig 1B). However, the protein has no N-terminal signal peptide32, suggesting that the amino terminus is located in the cytoplasm.
(A) Nucleotide and deduce amino acid sequences of Xenopus IU12 gene. The cDNA clone contains a 5′-UTR of 276 bases, followed by an open reading frame of 507 amino acids and a 461-base 3′-UTR. No concensus polyadenlylation signal is present in the 3′-UTR and a putative one is underlined (see text for discussions). The encoded polypeptide contains 12 transmembrane domains (in bold italics) (TM1-TM12) with the following order of likelihood to function as transmembrane segments (TM11>TM6> TM2>TM12> TM5>TM8> TM1 >TM7> TM10>TM9>TM3>TM4) (based on the analysis using the PSORT program at http://psort.nibb.ac.jp/form.html).
(B). Kyte-Doolittle hydrophobicity plot. The 12 predicted transmembrane domains are numbered as in Fig. 1A and are highly hydrophobic.
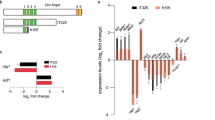
Sequence homology search showed that the predicted protein sequence is highly homologous to two proteins of unknown function but associated with cell activation (Fig 2). Although both the predicted human E16 and rat TA1 proteins are much smaller than IU12 (241 aa for E16 and TA1 vs 507 for IU12), the overlapped region is highly conserved and contains 6 of the 12 transmembrane domain of IU12 (Fig 2B). In addition, the IU12 polypeptide has over 82% identity (90% similarity) to both the human E16 protein33 and rat TA1 protein34. Even at the cDNA sequence level, there is over 73% identity among the three genes. Thus, IU12 is most likely the homolog of the human E16 and rat TA1.
Amino acid sequence comparison among Xenopus IU12, human E1633, and rat TA1[rTA1, 34]. Only the sequences that differ from IU12 are shown for hE16 and rTA1. Dots indicated the missing residues. Note that hE16 and rTA1 only contain the region corresponding to the carboxyl end 241 aa of the IU12 and thus only 6 of the 12 transmembrane domains (TM7-12) present in IU12. However, it is predicted that the full-length hE16 and rTA1 are likely to have all 12 TMs (see Discussion). In the overlapped region, the three proteins share over 82% identities and 90% similarities.
IU12 is regulated by T3 in selective organs
IU12 was originally isolated from a subtractive cDNA library made of intestinal mRNA from premetamorphic tadpoles treated with T3 for one day18. To determine whether IU12 can be induced by T3 in other organs, RNA was isolated from the intestine, brain, hindlimb, and tail of premetamorphic tadpoles at stages 52-54 that had been treated with or without T3 for one day. RNA was copied into cDNA using reverse transcripts, restricted to small fragments, ligated to a PCR-linker, and amplified by PCR. Southern blot hybridization was performed on these cDNAs by using IU12 as the probe (Fig 3). Varying levels of IU12 mRNA were detected in different tissues. There were little or no detectable mRNA in the intestine or hindlimb, and a very low level in the brain. T3 treatment led to a strong up-regulation of the mRNA levels in the intestine and limb but only a small increase in the brain. Finally, relative high levels of IU12 mRNA was present in the untreated tadpole tail and they were not changed by the hormone treatment. A hybridization with the control probe rpL8 indicated that rpL8 levels were independent of the treatment. Thus, the different levels of IU12 signals were not due to loading or PCR variations, and IU12 was responsive to T3 only in selective organs.
Organ-specific regulation of IU12 by T3. Tadpoles at stage 54 were treated in the presence (+) or absence (−) of T3 for up to 24 h. After treatment, poly (A)+ RNA was isolated from brain, hindlimb (limb), tail and intestine. RNA was copied into cDNA by reverse transcription, restricted to small fragments, ligated to a PCR-linker, and amplified by PCR. Southern blot hybridization was performed on the cDNA using an IU12 or rpL8 probe, which served as a control for loading and PCR. Note that the smeary signals were due to the procedure used to generate the PCR-cDNAs, which resulted in the restriction of full-length cDNAs into multiple fragments (see references in Materials and Methods). The variation in the migration of the cDNAs is due to the variation in the construction of the cDNA templates from mRNAs of different organs for PCR and subsequent PCR amplication. However, the blot serves the purpose to determine whether IU12 is regulated by T3 in different organs. It should be pointed out that two lines between the intestine and brain lanes were removed (for both the IU12 and rpL8 panels). This is because they contained cDNAs from the PCR amplification of cDNAs prepared from total RNAs not the poly A+ RNAs as in the rest of the blot.
Kinetic study of T3-induction in premetamorphic tadpole intestine demonstrated that the IU12 gene responded quickly to T3 treatment in the intestine with its mRNA detectable as early as 8 hr after the addition of T3 (Fig 4A). Similar kinetics has been observed for many other direct T3 response genes28, 29, 30, 31, including the Xenopus TRβ genes, which have been shown to be regulated at the transcription level by thyroid hormone receptors24. To determine whether IU12 by T3 is directly regulated by TRs, tadpoles were treated with or without T3 in the presence of protein synthesis inhibitors and RNA was isolated from the intestine for Northern blot analysis of the IU12 mRNA level (Fig 4B). The results showed that the IU12 mRNA was up-regulated both in the presence or absence of protein synthesis inhibitors. However, the protein synthesis inhibitors alone also increased the IU12 mRNA level (Fig 4B). Thus, the experiment failed to show whether IU12 regulation by T3 requires new protein synthesis, and direct analysis of the IU12 gene promoter will be needed to prove whether IU12 is a direct T3-response gene.
The IU12 gene is an early T3 response gene.
(A) Kinetics of T3 induction. Stage 54 tadpoles were treated with 5nM T3 and RNA was isolated from the intestine. Northern blot hybridization showed that the IU12 mRNA could be detected as early as 8 h after T3 addition. The positions of 28S and 18S rRNAs were indicated by the bars on the side.
(B) induction of IU12 gene in the presence of protein synthesis inhibitor. Stage 54 tadpoles were treated with 5nM T3 in the presence or absence of protein synthesis inhibitors (CHX) for 12 h. The RNA isolated and analyzed by Northern blot hybridization. Ten mg RNA was used per lane. The smeary signals migrated faster than the full length mRNA band were most likely due to partial degradations to the mRNA. The bars on the side indicate the positions of 18S and 28S rRNAs. The hybridization of the duplicate filters with rpL8 served as a loading control.
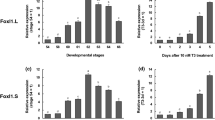
Developmental regulation of Xenopus IU12 gene
To investigate whether IU12 expression is regulated during metamorphosis, RNA was isolated from total tadpoles at different stages from premetamorphosis (stages 50-54) to the end of metamorphosis (stage 66) with or without one day T3 treatment. Northern blot analysis showed that IU12 was expressed throughout development (Fig 5). T3 treatment resulted in an increase in the IU12 mRNA level in premetamorphic tadpoles (stages 50-54) but had no effect on the mRNA level in stage 58 tadpoles. This was probably due the presence of high levels of endogenous T3 at stage 58 or later35, thus additional exogenous T3 would not significantly affect the animal development.
IU12 is regulated by exogenous T3 in premetamorphic tadpoles (stage 50-54) but independent of exogenous T3 during metamorphosis (stage 58-66). Tadpoles at different stages were treated with (+) or without (−) T3 and RNA was isolated from the whole tadpoles. Northern blot hybridization (10 μg per lane) was carried out with IU12 probe and the control rpL8 probe. All the lanes were from a single Nothern blot except that two lanes (between stage 58 and 62 lanes) containing RNA from the intestine alone were removed for easy comparisons among different stages.
As different organs undergo distinct transformations during metamorphosis. It is possible that IU12 expression may be regulated in an organ dependent manner. Thus, to study the possible role of IU12 gene in development, we chose the intestine as a model system. The intestine is known to undergo drastic transformation during metamorphosis6, 36. The tadpole intestine consists of mostly a simple tubular layer of larval epithelium with little connective tissue or muscle. During metamorphosis, the larval epithelium undergoes programmed cell death or apoptosis37, 38. Concurrently, the adult epithelial cells and the cells of the connective tissue and muscle proliferate and differentiate to form the adult intestine with a multiply folded adult epithelium39, 40.
When the IU12 gene expression was analyzed in the intestine of Xenopus tadpoles at different stages, its mRNA was found to be present in the intestine at stage 45 (Fig 6A) when tadpole feeding just began20. The expression was then repressed to lower levels in premetamorphic tadpoles. Beginning around stage 58, the IU12 mRNA levels were up-regulated as intestinal remodeling began6, 39, 40. The mRNA reached high levels by stage 62, the climax of metamorphosis when larval epithelial cell death was near the completion and adult epithelial cells were actively proliferating. The mRNA then remained at high levels in postmetamorphic frog intestine (stage 66). These results suggest that IU12 is involved in larval intestinal degeneration and adult intestinal organogenesis.
The Xenopus IU12 gene is highly expressed in the intestine during both natural and T3-induced metamorphosis. (A) Development expression of IU12 mRNA. Each lane had 10μg RNA. (B) IU12 expression during T3 treatment. Stage 54 tadpoles were treated with 5nM T3 for 0-6 d and RNA was isolated from the intestine. 5 μg RNA was used per land except the last lane which had only 2.5 μg (6 d treatments). The hybridization with rpL8 served as a loading control. The bars indicate the position of the 18S and 28S rRNA.
IU12 expression during T3-induced intestinal remodeling
Thyroid hormone is known to be able to induce precocious metamorphosis when added to the rearing water of premetamorphic tadpoles4, 5. If IU12 is involved in the remodeling of the intestine, its expression during T3-included metamorphosis should mimic that during natural development. Therefore, we treated stage 54 tadpoles for 0-6 d with 5 nM T3, a concentration similar to the maximal plasma T3 level at the climax of metamorphosis35. Total RNA was isolated from the intestine and IU12 expression was analyzed by Northern blot hybridization. Again, little IU12 mRNA was detected in control tadpole intestine and the IU12 mRNA was up-regulated after one day T3 treatment and reached a peak level in the intestine after two days of T3 treatment (Fig 6B). The IU12 gene continued to be expressed, although at varying levels, throughout the rest of the treatment. Under our treatment conditions, intestinal remodeling, including length reduction and epithelial folding22, is known to take place. The observed IU12 gene expression profile during the T3 treatment thus supports the suggestion that IU12 participates in adult intestinal development.
Discussion
We have reported here the cloning and expression studies of a T3-response gene isolated from the metamorphosing intestine. Our major findings are 1) that the gene IU12 encodes a homolog of a mammalian protein that is associated with cell activation and 2) that IU12 expression in the intestine implicates a role during the development and function of adult frog intestine.
Xenopus IU12 encodes a plasma membrane protein with 12 transmembrane domains
Northern blot analysis shows that the longest IU12 mRNA is about 5kb in length, considerately longer than the cDNA clone that we have isolated. However, some shorter transcripts were detected, especially those about 2-3 kb in length and migrating in an agarose gel between the 18 S and 28 S rRNAs. These could be due to partial degradation of the full length mRNA or due to alternative polyadenylation. Thus, the 2.3 kb cDNA clone that we have isolated may be one such alternatively polyadenylated mRNA. Its lack of a consensus polyadenylation signal may account of the relatively low abundance of the mRNA around 2.5 kb in length compared to the full-length mRNA. Another possibility is that the cDNA clone was derived from internal priming by dTn during cDNA synthesis for the construction of the cDNA library. Regardless of the exact origin of the cDNA clone, it encodes a full-length polypeptide of 507 aa with 12 predicted transmembrane domains but without a signal peptide for secretion32. This makes the protein most likely a plasma membrane protein with its N-terminus and C-terminus both located in the cytoplasm.
The predicted peptide sequence shares very high degrees of identities with the human E1633 and rat TA134, suggesting that IU12 is Xenopus homolog of the mammalian proteins. However, the mammation proteins are much shorter, containing only the region corresponding to the last 241 aa of the IU12 polypeptide. On the other hand, a DNA sequence comparison shows that the IU12 cDNA sequence shares high levels of conservation in a 250 bp (out of the 311 bp 5′-UTR) region with the human El6 gene and in a 70 bp (out of the 202 bp 5′-UTR) region with the rat gene (data not shown). Further more, there is no in-frame stop codon upstream of the first initiation codon in either TA1 or E16 cDNA sequences. Finally, the human E16 and rat TA1 cDNA clones are about 4 kb and 2.8 kb, while their mRNAs are about 4.8 kb and 3.2 kb, respectively33, 34. Since it is uncommon for the 5′-UTR to be highly conserved, it is very likely that the reported sequences for human E16 and rat TA1 cDNA represent partial mRNA sequences. The full length cDNAs for the human and rat gene may encode proteins with 12 transmembrane domains just like the Xenopus IU12 gene.
IU12 is likely a direct T3-response gene
Thyroid hormone is believed to regulate amphibian metamorphosis by inducing a cascade of gene expression2, 17. The immediate early or direct response genes refer to those whose transcription is under the direct control of T3 through TRs. Thus, their regulation by T3 is expected to be fast and independent of new protein synthesis. The activation of IU12 is within the early reponse gene class18, 29, with its mRNA levels up-regulated within 4-8 h after the addition of T3 to the rearing water of premetamorphic tadpoles. However, due to the up-regulation of its mRNA level by the protein synthesis inhibitors, we failed to determine whether the induction of IU12 by T3 requires new protein synthesis. On the other hand, several T3-reponse genes with similar induction kinetics, eg. TRβ, stromelysin-3, and hedgehog genes18, 28, 29, have been shown to be up-regulated even in the presence of protein synthesis inhibitors. Furthermore, the Xenopus TRβ genes have been demonstrated to be directly regulated by TRs themselves24. Thus, it is quite likely that IU12 is also a direct T3-reponse gene.
A role of IU12 gene in intestinal remodeling and adult intestinal function
The IU12 gene has very little expression in premetamorphic intestine. The gene begins to be up-regulated around stage 58 in the intestine just at the beginning of intestinal metamorphosis. Similarly, during T3 treatment of premetamorphic tadpoles, IU12 mRNA is up-regulated within one day and reaches a peak level after two days, prior to visible cell death in the intestine19, 22. This suggest that IU12 is involved in the degeneration of larval intestinal epithelium through programmed cell death. In addition, IU12 mRNA is abundantly expressed in the intestine at the climax of metamorphosis (stage 62), when active proliferation of the adult epithelial cells and cells of the connective tissue and muscles takes place. These high levels of expression persists in postmetamorphic frogs. Furthermore, IU12 expression, once induced, also persists throughout the T3 treatment of tadpoles. These results argue for a role of IU12 in the formation of the adult intestine and maintenance of adult intestinal function in frogs.
How IU12 participates in intestinal remodeling remains to be determined. As a transmembrane protein, it may be involved in certain signal translation pathways that are associated with cell proliferation and/or cell death. In this regard, it is interesting to note that the human homolog E16 is associated with cell activation33 and that the rat homolog TA1 is also associated with cell activation as well as liver development and carcinogenesis34. Thus, in addition to sequence homologies, these vertebrate transmembrane proteins also share conserved functions in cell proliferation and organ development. The expression of Xenopus IU12 during early stage of intestinal remodeling also implicates that if IU12 is expression in differentiated cell types such as the intestinal epithelial cells, it may facilitate cell death. Future studies on in situ localization of IU12-expressing cells and functional characterization should be able to test these hypotheses directly.
Accession codes
References
Atkinson BG . Metamorphosis: Model systems for studying gene expression in postembryonic development. Developmental Genet 1994; 15:313–9.
Shi Y-B . Molecular biology of amphibian metamorphosis: A new approach to an old problem. Trends Endocrinol Metab 1994; 5:14–20.
Tata JR . Gene expression during metamorphosis: An ideal model for post-embryonic development. BioEssays 1993; 15:239–48.
Dodd MHI, Dodd JM . The biology of metamorphosis. In physiology of the amphibia(ed. B.Lofts), pp. 467–599. Academic Press, New York. 1976.
Gilbert LI, Frieden E . Metamorphosis: A problem in developmental biology, 2nd ed., Plenum Press, New York. 1981.
Shi Y-B, Ishizuya-Oka A . Biphasic intestinal development in amphibians: Embryogensis and remodeling during metamorphosis. Current Topics in Develop Biol 1996; 32:205–35.
Balls M, Bownes M . Metamorphosis. The Clarendon Press, Oxford, UK. 1985.
Yoshizato K . Biochemistry and cell biology of amphibian metamorphosis with a special emphasis on the mechanism of removal of larval organs. Inst Rev Cytol 1989; 119:97–149.
Galton VA . Thyroid hormone action in amphibian metamorphosis. In Oppenheimer, J.H., Samuels, H.H., eds. Molecular basis of thyroid hormone action. New York, Academic, 1983; pp 445–83.
Kikuyama S, Kawamura K, Tanaka S, Yamamoto K . Aspects of amphibian metamorphosis: hormonal control. Int Rev Cytol 1993; 145:105–48.
Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM . The nuclear receptor superfamily: The second decade. Cell 1995; 83:835–9.
Sap J, Munoz A, Damm K, Goldberg Y, Ghysdael J, Leutz A, Berg H, Vennstrom B . The C-erb-A protein is a high affinity receptor for thyroid hormone. Nature 1986; 324:635–40.
Shi YB, Wong J, Puzianowska-Kuznicka M . Thyroid hormone receptors: Mechanisms of transcriptional regulation and roles during frog development. J Biomed Sci 1996; 3:307–18.
Tsai M-J, O'Malley BW : Molecular mechanisms of action of steroid/thyroid receptor receptor superfamily members. Ann Rev Biochem 1994; 63:451–86.
Weinberger C, Thompson CC, Ong ES, Lebo R, Gruol D J, Evans RM . The c-erb-A gene encodes a thyroid hormone receptor. Nature 1986; 324:641–6.
Brown DD, Wang Z, Furlow JD, Kanamori A, Schwartzman RA, Remo BF, Pinder A . The thyroid hormone-induced tail resorption program during Xenopus laevis metamorphosis. PNAS 1996; 93:1924–9.
Gilbert LI, Tata JR, Atkinson BG . Metamorphosis: Post-embryonic reprogramming of gene expression in amphibian and insect cells. Academic Press, New York 1996.
Shi Y-B, Brown DD . The earliest changes in gene expression in tadpole intestine induced by thyroid hormone. J Biol Chem 1993; 266:20312–7.
Patterton D, Hayes WP, Shi Y-B . Transcriptional activation of the matrix metaoproteinase gene stromelysin-3 coincides with thyroid hormone-induced cell death during frog metamorphosis. Dev Biol 1995; 167:252–62.
Nieuwkoop PD, Faber J . Normal table of Xenopus laevis. North Holland Publishing, Amsterdam. 1956.
Kanamori A . Brown DD . The regulation of thyroid hormone receptor b genes by thyroid hormone in Xenopus laevis. J. Biol. Chem. 1992; 267:739–45.
Shi Y-B, Hayes WP . Thyroid hormone-dependent regulation of the intestinal fatty acid-binding protein gene during amphibian metamorphosis. Dev Biol 1994; 161:48–58.
Maniatis T, Fritsch EF, Sambrook J . Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York 1982.
Ranjan M, Wong J, Shi Y-B . Transcriptional repression of Xenopuf TRβ gene is mediated by a thyroid hormone response element located near the start site. The Journal of Biol Chem 1994; 269:24699–705.
Herrin DL, Schmidt GW . Rapid reversible staining of Northern blots prior to hybridization. BioTechniques 1988; 6:196–200.
Shi Y-B, Liang VC-T . Cloning and characterization of the ribosomal protein L8 gene from Xenopus laevis. Biochimica et Biophysica Acta. 1994; 1217:227–8.
Buckbinder L, Brown DD . Thyroid hormone-induced gene expression changes in the developing frog limb. J Biol Chem 1992; 267:25786–91.
Stolow MA, Shi Y-B . Xenopus sonic hedgehog as a potential morphogen during embryogenesis and thyroid hormone-dependent metamorphosis. Nucl Acids Res 1995; 23:2555–62.
Wang Z, Brown DD . Thyroid hormone-induced gene expression program for amphibai tail resorption. J Biol Chem 1993; 268:16270–78.
Ishizuya-Oka A, Stolow MA, Ueda S, Shi Y-B . Temporal and Spatial Expression of An Intestinal Na+/PO43− Cotransporter Correlates With Epithelial Transformation During Thyroid Hormone-Dependent Frog Metamorphosis. Dev Gen 1997; 20:53–66.
Patterton D, Shi Y-B . Thyroid Hormone-dependent Differential Regulation of Multiple Arginase Genes during Amphibian Metamorphosis. J Biol Chem 1994; 269:25328–34.
Von Heijne G . A new method for predicting sigmal sequence cleavage sites. Nucleic Acids Research 1986; 14:4683–90.
Gaugitsch HW, Prieschl EE, Kalthoff F, Huber NE, Baumruker TA . Novel Transiently Expressed, Integral Membrane Protein Linked to Cell Activation. J Biol Chem 1992; 267:11267–73.
Sang J, Lim YP, Panziaca M, Finch P, Thomspon NL . TA1, a highly conserved oncofetal complementary DNA from rat hepatoma, encodes an integral membrane protein associated with liver development, carcinogenesis, and cell activation. Cancer Res 1995; 55:1152–9.
Leloup J, Buscaglia M . La triiodothyronine: hormone de la mtamorphose des amphibiens. CR Acad Sci 1977; 284:2261–3.
Dauca M . Hourdry J . Transformations in the intestinal epithelium during anuran metamorphosis. In Metamorphosis (Balls M. and Bownes, M., eds). pp. 36–58. The clarendon Press, Oxford, UK. 1985.
Ishizuya-Oka A, and Ueda S . Apoptosis and cell proliferation in the Xenopus small intestine during metamorphosis. Cell Tissue Res 1996; 286:467–76.
Su Y, Shi Y, Shi Y-B . Cyclosporin A but not FK 506 inhibits thyroid hormoneinduced apoptosis in tadpole intestinal epithelium. FASEB J 1997; 11:559–65.
Ishizuya-Oka A, Shimozawa A . Development of the connective tissue in the digestive tract of the larval and metamorphosing Xenopus laevis. Ant Anz Jena 1987; 164:81–93.
McAvoy JW, Dixon KE . Cell proliferation and renewal in the small intestinal epithelium of metamorphosing and adult Xenopus laevis. J Exp Zool 1977; 202:129–38.
Acknowledgements
We would like to thank Ms. Kieu Pham for preparing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liang, V., Sedgwick, T. & Shi, Y. Characterization of the Xenopus homolog of an immediate early gene associated with cell activation: sequence analysis and regulation of its expression by thyroid hormone during amphibian metamorphosis. Cell Res 7, 179–193 (1997). https://doi.org/10.1038/cr.1997.19
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/cr.1997.19