Abstract
Simultaneous genomic in situ hybridization (GISH) with probe preannealing was used to detect the relationship between chromosomal position and sequence conservation on Aegilops speltoides var. aucheri chromosomes. DNA of Secale sylvestre, Hordeum spontaneum, Festuca pratensis, Semiarundinaria fastuosa, Arundo donax and Zea mays that represent several main groups of Poaceae were used as probes. Different GISH-banding patterns that characterize diverse evolutionary trajectories in the repetitive DNA fraction and correlate with evolutionary distance between tested species were observed. Fast-evolving sequences were detected in subterminal telomeric and subtelomeric heterochromatic regions, whereas sequences in pericentromeric regions showed high levels of conservation. GISH experiments revealed extensive conservation in NOR regions on chromosomes 1 and 6 which, in fact, appears to be a complicated mix of rDNA clusters and heterochromatin blocks of different nucleotide composition.
Similar content being viewed by others
Introduction
The method of genomic in situ hybridization (GISH), with total genomic DNA as a probe in hybridization experiments, provides unique information about similarities between repetitive DNAs from related species, as well as the physical location of conserved sequences on chromosomes (Fuchs et al., 1996; Heslop-Harrison & Schwarzacher, 1996). Simultaneous GISH with probe preannealing permits differences at the chromosome level to be displayed between chromatin fractions of a model species (Belyayev & Raskina, 1998). This type of data could shed light on the function and evolutionary dynamics of the noncoding DNA fraction. It is known that different chromosomal segments have different levels of sequence conservation. There are several chromosome structures that can be detected at the cytogenetical level such as rDNA loci or terminal telomeric sequences that share much in common with those from animals and fungi, hence there are some minor changes over a billion years (Appels & Honeycutt, 1986; Kipling, 1995). On the other hand, some chromosome regions show rapid changes in comparatively short time periods and some differences can be detected even at the intraspecies level (Kipling, 1995; Belyayev & Raskina, 1998).
In the present study we aimed to identify evolutionary dynamics and chromosomal distribution of repetitive DNA by hybridizing Aegilops speltoides Tausch chromosomes with DNA of species representing several groups of Poaceae using the GISH technique. Ae. speltoides belongs to the section Sitopsis (Triticeae) and is most similar to the donor of the B-genome of bread wheat (Maestra & Naranjo, 2000).
Specifically, we investigated two questions. (1) In what order are chromosomal segments involved in the process of evolutionary divergence, i.e. what is the relationship between chromosomal position and sequence conservation? (2) At what evolutionary distance is it still possible to find cytologically visible similarity in repetitive noncoding DNA sequences between Poaceae species?
Materials and methods
Aegilops speltoides var. aucheri (2n=2x=14) from Latakia Province, Syria, original germplasm source, USDA PI 487235, was used for preparations of chromosome spreads and DNA isolation. DNA was also obtained from the following species: Secale sylvestre Host., Russia, Komarov Botanical Institute collection; Hordeum spontaneum C. Koch, from Mt. Hermon, Israel, Institute of Evolution collection; Festuca pratensis Huds., Russia, Komarov Botanical Institute collection; Semiarundinaria fastuosa (Mitf.) Makino ex Nakai, Russia, Komarov Botanical Institute collection; Arundo donax L., from nature, near Nes-Tsiona, Israel; Zea mays L., Weizmann Institute collection; Lycopersicon esculentum L, Weizmann Institute collection. These species (except L. esculentum) represent several evolutionary branches of the Poaceae (Fig. 1) S. sylvestre is closely related to Ae. speltoides and belongs to the same subtribe Triticinae, whereas Z. mays and A. donax are distantly related. The dicotyledon L. esculentum was selected to discriminate highly conserved chromosome regions.
Scheme of evolutionary separation between investigated species. Dendrogram is according to Tsvelev (1976). In the text boxes approximate time of taxa divergence million years ago (mya) are shown.
GISH procedure was described previously in detail (Belyayev & Raskina, 1998; Belyayev et al., 2000). Sonicated genomic DNA from Aegilops speltoides was labelled with biotin and preannealed with digoxigenin-labelled sonicated total genomic DNA from each of the other species at 37°C overnight. The probe mixture was hybridized to mitotic chromosomes of Ae. speltoides. The detection of biotin with fluorescein isothiocyanate (FITC)-conjugated avidin (green fluorescence) and digoxigenin with rhodamine-conjugated sheep antidigoxigenin Fab fragment (red fluorescence) was made simultaneously. As a result, chromosome regions of preferential hybridization of self-DNA were visualized as green, whereas regions of combined hybridization showed orange-yellow fluorescence.
We established several steps for standardization of experimental procedures to be sure that data were comparable and reproducible. Several preliminary experiments to optimize the GISH protocol with regard to optimal concentrations, temperatures and other variables were made. Only the optimized GISH protocol was used for experiments. Previously (Belyayev & Raskina, 1998), we carried out a control experiment in which equal amounts of DNA from Ae. speltoides were separately labelled with biotin and digoxigenin, mixed in a 1:1 ratio and, following overnight preannealing, were hybridized to Ae. speltoides chromosomes. No green bands were detected on the yellow-orange chromosomes, hence both probes hybridized evenly along the chromosomes.
We repeated each experiment three times (two slides in each hybridization). In each slide we analysed a minimum of five complete metaphase plates. Chromosomes were identified according to Friebe & Gill (1996) as described in Belyayev & Raskina (1998).
Results
1 GISH using DNA from Ae. speltoides and Secale sylvestre on the chromosomes of Ae. speltoides. We obtained an orange-yellow combined signal that resulted from hybridization of both Ae. speltoides (detected in green) and S. sylvestre (detected in red) DNA in pericentromeric and intercalary chromosome regions. Green bands resulted from preferential hybridization of Ae. speltoides DNA to subterminal telomeric (all chromosomes) and subtelomeric (long arms of chromosomes 2, 3, 4, 5 and 7) heterochromatic regions (Figs 2A and 3).
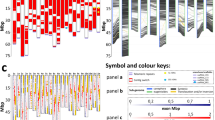
GISH on the chromosomes of Aegilops speltoides. A. Probe: DNA of Ae. speltoides and Secale sylvestre. B. Probe: DNA of Ae. speltoides and Hordeum spontaneum. C. Probe: DNA of Ae. speltoides and Festuca pratensis. D. Probe: DNA of Ae. speltoides and Semiarundinaria fastuosa. Pro-metaphase plate. E. Probe: DNA of Ae. speltoides and Semiarundinaria fastuosa. Metaphase plate. F. DNA of Ae. speltoides and Zea mays. G. Probe: DNA of Ae. speltoides and Lycopersicon esculentum (Solanaceae). Bar 10 μm. Signals from rRNA gene locations on Ae. speltoides chromosomes 1 and 6 are arrowed in D–G.
Summarized GISH-banding karyotype of Ae. speltoides (left part of chromosome) versus C-banding (right part of chromosome). Heterochromatin discriminated according to its conservation. Sequences of these regions are different between Ae. speltoides and: red — S. sylvestre; dark red — H. spontaneum; green — F. pratensis; dark green — A. donax. Highly conserved regions that are common for all investigated species including Z. mays and S. fastuosa are shown in blue.
2 GISH using DNA from Ae. speltoides and Hordeum spontaneum on the chromosomes of Ae. speltoides. DNA of H. spontaneum hybridized mainly to pericentromeric regions and in the regions of secondary constrictions (Figs 2B and 3). On chromosomes 1, 4, 5 and 6 the hybridization sites correspond to the pericentromeric heterochromatin regions revealed by Hoechst-33258 staining. On chromosomes 2, 3 and 7 the size of the hybridization sites exceeded that of the heterochromatic regions. In euchromatic regions a dispersed red signal of low intensity was detected. Telomeric and subtelomeric heterochromatin regions showed green fluorescence of high intensity. These data are similar to those obtained previously (Belyayev & Raskina, 1998).
3 GISH using DNA from Ae. speltoides and Festuca pratensis on the chromosomes of Ae. speltoides. Compared with the previous experiment, there was a further decrease of total red signal (Figs 2C and 3). DNA of F. pratensis also hybridized mainly in pericentromeric regions, but the sizes of these bands were reduced. Hybridization also revealed orange bands in the NOR regions.
4 GISH using DNA from Ae. speltoides, Arundo donax, Zea mays and Semiarundinaria fastuosa on the chromosomes of Ae. speltoides. Most distinct signals from hybridization of DNA of these species appeared in the NOR regions on chromosomes 1 and 6. An orange signal of low intensity was also observed in some pericentromeric regions (Figs 2D, E, F and 3).
5 GISH using DNA from Ae. speltoides and Lycopersicon esculentum on the chromosomes of Ae. speltoides. In this experiment, orange signal of low intensity from the DNA of L. esculentum could be observed only in NOR regions (Fig. 2G).
Discussion
In cereals, the amount of repetitive DNA families that serve as a base for genomic in situ hybridization reaches up to 85% of the total DNA (Flavell et al., 1993; Heslop-Harrison & Schwarzacher, 1996). Using the GISH technique with probe preannealing (to increase probe specificity) we obtained several types of banding patterns on chromosomes of Ae. speltoides. The high temperature of hybridization allows high-precision annealing of chromosome/probe sequences (Bostock & Sumner, 1978), and at the same time it sharpens differences between reassociation kinetics of eu- and heterochromatin, revealing chromosome substructure. The hybridization mixture consists of self-DNA of Ae. speltoides detected in green and DNA of one of the tested species detected in red (probes overlapping). Thus, the green signal shows chromosome regions with preferential hybridization of Ae. speltoides DNA. The combined signal that is visualized as red–orange fluorescence shows regions of hybridization of both DNAs, i.e. the chromosomal distribution of common sequences. The alignment of these signals shows correlation with evolutionary distance (Fig. 1) (Tsvelev, 1976). By comparison of the patterns obtained it is possible to reconstruct the dynamics of the evolutionary processes in repetitive DNA families in the whole genome and in single chromosomes (Figs 2 and 3).
The green hybridization pattern that was obtained in the experiment with DNA of S. sylvestre from the same subtribe Triticinae (Tsvelev, 1976), shows the physical location on chromosomes of Ae. speltoides of rapidly evolving genus-specific repetitive sequences (Fig. 2A, red in Fig. 3). It corresponds to the subterminal telomeric and part of the subtelomeric heterochromatin regions (Belyayev & Raskina, 1998). Sequences in these regions are therefore the first to be involved in the process of evolutionary divergence in Ae. speltoides. This correlates with the known fact that in many species terminal repeat arrays and unique internal sequences are separated by repetitive sequences which are often species-specific, and many of which are found only at telomeres (Anamthawat-Jonsson & Heslop-Harrison, 1993; Kipling, 1995). The origin of the species-specific sequences is unclear. This can be due to accumulation of mutations caused by different factors, by activity of mobile elements, for instance (Belyayev et al., 2001), and/or due to replacement of sequences by other classes of repeats.
Probing chromosomes of Ae. speltoides with DNA of H. spontaneum (subtribe Hordeinae) revealed a decrease of total red signal as compared with the previous experiment (Fig. 2B, dark red in Fig. 3). The red signal of low intensity can still be observed in intercalar hetero-and euchromatin regions, but the main red pattern corresponds to pericentromeric regions. This GISH experiment and hybridization with the DNA of a species that does not belong to the tribe Triticeae reveals pericentromeric heterochromatin as the location of slow-evolving family-specific sequences (Figs 2C, D and 3). It has long been established that the centromere suppresses the processes of recombination and unequal crossing over (Choo, 1997) that appear to be one of the main evolutionary mechanisms (Elder & Turner, 1995). Several repetitive DNA elements were found in centromeric regions through all cereals (Aragón-Alcaide et al., 1996; Dong et al., 1998). Most likely, this evolutionary conservation results from the centromere’s functional importance.
Hybridization of Ae. speltoides chromosomes with DNA of species that do not belong to the subfamily Pooideae (Fig. 1) (Tsvelev, 1976) – S. fastuosa (Bambusoideae) (Fig. 2D, E, from dark green to blue on Fig. 3), A. donax (Arundinoideae) (data not shown) and Z. mays (Panicoideae) (Fig. 2F) gave very similar results. Red signal of low intensity was observed in pericentromeric regions, most distinctly on prometaphase chromosomes (Fig. 2D). The size of these regions does not exceed that revealed by Hoechst-33258 staining (Belyayev & Raskina, 1998). Bright orange bands of combined hybridization of self and alien DNA shows sequence similarity in NOR regions on chromosomes 1 and 6 which, in fact, appears to be a complicated mix of rDNA clusters (Badaeva et al., 1996) and heterochromatin blocks of different nucleotide composition (Schweizer, 1980; Belyayev & Raskina, 1998). The signal from those regions was also observed when DNA from the dicotyledon L. esculentum (Fig. 2F) was used.
Our data obtained from GISH experiments on Ae. speltoides chromosomes show that the first evolutionary changes that can be detected at the chromosome level in noncoding repetitive DNA sequences take place in subterminal telomeric heterochromatin. Cytologically visible similarity in noncoding repetitive DNA sequences is still detectable at pericentromeric regions of Poaceae species belonging to different tribes.
References
Anamthawat-Jonsson, K. and Heslop-Harrison, J. S. (1993). Isolation and characterization of genome-specific DNA sequences in Triticeae species. Mol Gen Genet, 240: 151–158.
Appels, R. and Honeycutt, R. L. (1986). rDNA: evolution over a billion years. In: Dutta, S. K. (ed.) DNA Systematics: Plants, pp. 81–125. CRC Press, Boca Raton.
Aragón-Alcaide, L., Miller, T., Schwarzacher, T., Reader, S. et al. (1996). A cereal centromere sequence. Chromosoma, 105: 61–268.
Badaeva, E. D., Friebe, B. and Gill, B. S. (1996). Genome differentiation in Aegilops. 2. Physical mapping of 5S and 18S–26S ribosomal RNA gene families in diploid species. Genome, 39: 1150–1158.
Belyayev, A. and Raskina, O. (1998). Heterochromatin discrimination in Aegilops speltoides by simultaneous genomic in situ hybridization. Chromosome Res, 6: 559–565.
Belyayev, A., Raskina, O., Korol, A. and Nevo, E. (2000). Coevolution of A and B-genomes in allotetraploid Triticum dicoccoides. Genome, 43: 1021–1026.
Belyayev, A., Raskina, O. and Nevo, E. (2001). Chromosomal distribution of reverse transcriptase-containing retroelements in two Triticeae species. Chromosome Res, 9: 129–136.
Bostock, J. C. and Sumner, A. T. (1978). The Eucaryotic Chromosome. North-Holland Publishing Company, Amsterdam, New York, Oxford.
Choo, K. H. A. (1997). The Centromere. Oxford University Press, Oxford, New York, Tokyo.
Dong, F., Miller, J. T., Jackson, S. A., Wang, G. -L. et al. (1998). Rice (Oryza sativa) centromeric regions consist of complex DNA. Proc Nat Acad Sci USA, 95: 8135–8140.
Elder, J. F. and Turner, B. J. (1995). Concerted evolution of repetitive DNA sequences in eucaryotes. Q Rev Biol, 70: 297–320.
Flavell, R. B., Gale, M., O'Dell, M., Murphy, G. et al. (1993). Molecular organization of genes and repeats in large cereal genomes and implications for the isolation of genes by chromosome walking. Chromosomes Today, 11: 199–214.
Friebe, B. and Gill, B. S. (1996). Chromosome banding and genome analysis in diploid and cultivated polyploid wheats. In: Jauhar, P. P. (ed) Methods of Genome Analysis in Plants. CRC Press, Boca Raton, NY. pp. 39–60.
Fuchs, J., Houben, A., Brandes, A. and Schubert, I. (1996). Chromosome ‘painting’ in plants – a feasible technique? Chromosoma, 104: 315–320.
Heslop Harrison, J. S. and Schwarzacher, T. (1996). Genomic Southern and In Situ hybridization for plant genome analysis. In: Jauhar, P. P. (ed.) Methods of Genome Analysis in Plants. CRC Press, Boca Raton, NY. 163–179.
Kipling, D. (1995). The Telomere. Oxford University Press, Oxford, New York, Tokyo.
Maestra, B. and Naranjo, T. (2000). Genome evolution of Triticeae. Chromosomes Today, 13: 155–167.
Schweizer, D. (1980). Fluorescent chromosome banding in plants: applications, mechanisms, and implications for chromosome structure. In: Davies, D. R. and Hopwood, R. A. (eds) Proc. 4th John Innes Symposium. Norwich 1979: The Plant Genome, pp. 61–72. John Innes Charity, Norwich.
Tsvelev, N. N. (1976). Cereals of USSR. Nauka, Leningrad (in Russian).
Acknowledgements
The authors are most grateful to Professor Jonathan F. Wendel, Iowa State University, for helpful discussions. We thank Professor Abraham Levi, Weizmann Institute, and Dr Violleta Kotseruba, Komarov Botanical Institute, for providing material for the research.
A.B. and O.R. thank the Ministry of Absorption of Israel for the supporting of this research. E.N. is indebted to the Israel Discont Bank Chair of Evolutionary Biology, and the Ancell–Teicher Research Foundation for Genetics and Molecular Evolution.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Belyayev, A., Raskina, O. & Nevo, E. Evolutionary dynamics and chromosomal distribution of repetitive sequences on chromosomes of Aegilops speltoides revealed by genomic in situ hybridization. Heredity 86, 738–742 (2001). https://doi.org/10.1038/sj.hdy.6888910
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6888910
Keywords
This article is cited by
-
Genomic constitution, allopolyploidy, and evolutionary proposal for Cynodon Rich. based on GISH
Protoplasma (2022)
-
Chromosomal structures and repetitive sequences divergence in Cucumis species revealed by comparative cytogenetic mapping
BMC Genomics (2015)
-
Constructing an alternative wheat karyotype using barley genomic DNA
Journal of Applied Genetics (2015)
-
Isolation and characterization of reverse transcriptase fragments of LTR retrotransposons from the genome of Chenopodium quinoa (Amaranthaceae)
Plant Cell Reports (2013)
-
Identification of LMW Glutenin-Like Genes from Secale sylvestre Host
Russian Journal of Genetics (2005)