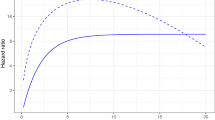
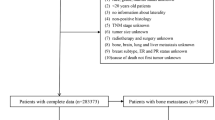
Abstract
Accurate prediction of the individualized survival benefit of adjuvant therapy is key to making informed therapeutic decisions for patients with early invasive breast cancer. Machine learning technologies can enable accurate prognostication of patient outcomes under different treatment options by modelling complex interactions between risk factors in a data-driven fashion. Here, we use an automated and interpretable machine learning algorithm to develop a breast cancer prognostication and treatment benefit prediction model—Adjutorium—using data from large-scale cohorts of nearly one million women captured in the national cancer registries of the United Kingdom and the United States. We trained and internally validated the Adjutorium model on 395,862 patients from the UK National Cancer Registration and Analysis Service (NCRAS), and then externally validated the model among 571,635 patients from the US Surveillance, Epidemiology, and End Results (SEER) programme. Adjutorium exhibited significantly improved accuracy compared to the major prognostic tool in current clinical use (PREDICT v2.1) in both internal and external validation. Importantly, our model substantially improved accuracy in specific subgroups known to be under-served by existing models. Adjutorium is currently implemented as a web-based decision support tool (https://vanderschaar-lab.com/adjutorium/) to aid decisions on adjuvant therapy in women with early breast cancer, and can be publicly accessed by patients and clinicians worldwide.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The dataset used to derive and internally validate the model was obtained from the National Cancer Registration and Analysis Service. These data are held by Public Health England. Information on how to access the data is available at http://ncin.org.uk/collecting_and_using_data/data_access. The dataset used for external validation was obtained from the Surveillance, Epidemiology and End Results programme, which can be accessed at https://seer.cancer.gov/seertrack/data/request/.
Code availability
The code for the AutoPrognosis software is available at https://bitbucket.org/mvdschaar/mlforhealthlabpub.
References
Fitzmaurice, C. et al. Global, regional and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 3, 524–548 (2017).
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018).
Guo, F., Kuo, Y.-f, Shih, Y. C. T., Giordano, S. H. & Berenson, A. B. Trends in breast cancer mortality by stage at diagnosis among young women in the United States. Cancer 124, 3500–3509 (2018).
Sparano, J. A. et al. Clinical and genomic risk to guide the use of adjuvant therapy for breast cancer. New Engl. J. Med. 380, 2395–2405 (2019).
Symmans, W. F. et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J. Clin. Oncol. 25, 4414–4422 (2007).
Wishart, G. C. et al. PREDICT: a new UK prognostic model that predicts survival following surgery for invasive breast cancer. Breast Cancer Res. 12, R1 (2010).
dos Reis, F. J. C. et al. An updated predict breast cancer prognostication and treatment benefit prediction model with independent validation. Breast Cancer Res. 19, 58 (2017).
Shachar, S. S. & Muss, H. B. Internet tools to enhance breast cancer care. NPJ Breast Cancer 2, 16011 (2016).
Kattan, M. W. et al. American joint committee on cancer acceptance criteria for inclusion of risk models for individualized prognosis in the practice of precision medicine. CA Cancer J. Clin. 66, 370–374 (2016).
Early and Locally Advanced Breast Cancer: Diagnosis and Management, NICE Guideline NG101 (National Institute for Health and Care Excellence, 2018).
van Maaren, M. C. et al. Validation of the online prediction tool PREDICT v.2.0 in the Dutch breast cancer population. Eur. J. Cancer 86, 364–372 (2017).
Olivotto, I. A. et al. Population-based validation of the prognostic model ADJUVANT! for early breast cancer. J. Clin. Oncol. 23, 2716–2725 (2005).
Bhoo-Pathy, N. et al. ADJUVANT! Online is overoptimistic in predicting survival of Asian breast cancer patients. Eur. J. Cancer 48, 982–989 (2012).
Campbell, H., Taylor, M., Harris, A. & Gray, A. An investigation into the performance of the ADJUVANT! Online prognostic programme in early breast cancer for a cohort of patients in the United Kingdom. Br. J. Cancer 101, 1074–1084 (2009).
Miao, H. et al. Validation of the CancerMath prognostic tool for breast cancer in Southeast Asia. BMC Cancer 16, 820 (2016).
Ravdin, P. M. et al. Computer program to assist in making decisions about adjuvant therapy for women with early breast cancer. J. Clin. Oncol. 19, 980–991 (2001).
Obermeyer, Z. & Emanuel, E. J. Predicting the future-big data, machine learning and clinical medicine. New Engl. J. Med. 375, 1216–1219 (2016).
Chen, J. H. & Asch, S. M. Machine learning and prediction in medicine-beyond the peak of inflated expectations. New Engl. J. Med. 376, 2507–2509 (2017).
Alaa, A. & Schaar, M. AutoPrognosis: automated clinical prognostic modeling via Bayesian optimization with structured kernel learning. In Proc. 35th International Conference on Machine Learning Vol. 80, 139–148 (PMLR, 2018).
Alaa, A. M. & van der Schaar, M. Demystifying black-box models with symbolic metamodels. In Advances in Neural Information Processing Systems 11301–11311 (NIPS, 2019).
Early Breast Cancer Trialists Collaborative Group Comparisons between different polychemotherapy regimens for early breast cancer: meta-analyses of long-term outcome among 100,000 women in 123 randomised trials. Lancet 379, 432–444 (2012).
Romond, E. H. et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. New Engl. J. Med. 353, 1673–1684 (2005).
Alaa, A. M. & van der Schaar, M. Prognostication and risk factors for cystic fibrosis via automated machine learning. Sci. Rep. 8, 11242 (2018).
Alaa, A. M., Bolton, T., Di Angelantonio, E., Rudd, J. H. & van Der Schaar, M. Cardiovascular disease risk prediction using automated machine learning: a prospective study of 423,604 UK biobank participants. PLoS ONE 14, e0213653 (2019).
Lambert, J. & Chevret, S. Summary measure of discrimination in survival models based on cumulative/dynamic time-dependent ROC curves. Stat. Methods Med. Res. 25, 2088–2102 (2016).
Harrell, F. E.Jr, Lee, K. L. & Mark, D. B. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat. Med. 15, 361–387 (1996).
Uno, H., Cai, T., Pencina, M. J., D’Agostino, R. B. & Wei, L. On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat. Med. 30, 1105–1117 (2011).
Noone, A. et al. SEER Cancer Statistics Review, 1975–2015 (National Cancer Institute, 2018).
Galea, M. H., Blamey, R. W., Elston, C. E. & Ellis, I. O. The Nottingham Prognostic Index in primary breast cancer. Breast Cancer Res. Treat. 22, 207–219 (1992).
Michaelson, J. S. et al. Improved web-based calculators for predicting breast carcinoma outcomes. Breast Cancer Res. Treat. 128, 827–835 (2011).
Zhang, Z. Multiple imputation with multivariate imputation by chained equation (MICE) package. Ann. Transl. Med. 4, 30 (2016).
Kotsiantis, S. B., Zaharakis, I. & Pintelas, P. Supervised machine learning: a review of classification techniques. Emerg. Artif. Intell. Appl. Comput. Eng. 160, 3–24 (2007).
Yersal, O. & Barutca, S. Biological subtypes of breast cancer: prognostic and therapeutic implications. World J. Clin. Oncol. 5, 412–424 (2014).
Down, S. K., Lucas, O., Benson, J. R. & Wishart, G. C. Effect of PREDICT on chemotherapy/trastuzumab recommendations in HER2-positive patients with early-stage breast cancer. Oncol. Lett. 8, 2757–2761 (2014).
Wishart, G. C. et al. Inclusion of KI67 significantly improves performance of the PREDICT prognostication and prediction model for early breast cancer. BMC Cancer 14, 908 (2014).
Ács, B. et al. Ki-67 as a controversial predictive and prognostic marker in breast cancer patients treated with neoadjuvant chemotherapy. Diagn. Pathol. 12, 20 (2017).
Ware, J. H., Harrington, D., Hunter, D. J. & D’Agostino, R. B.Sr Missing data. New Engl. J. Med. 367, 1353–1354 (2012).
Royston, P. Multiple imputation of missing values. Stata J. 4, 227–241 (2004).
Topol, E. J. High-performance medicine: the convergence of human and artificial intelligence. Nat. Med. 25, 44–56 (2019).
Latimer, N., Abrams, K. & Siebert, U. Two-stage estimation to adjust for treatment switching in randomised trials: a simulation study investigating the use of inverse probability weighting instead of re-censoring. BMC Med. Res. Methodol. 19, 69 (2019).
Mayer, E. L. Targeting breast cancer with CDK inhibitors. Curr. Oncol. Rep. 17, 443 (2015).
D’Agostino, R. & Nam, B.-H. Evaluation of the performance of survival analysis models: discrimination and calibration measures. Handbook Stat. 23, 1–25 (2003).
Acknowledgements
We thank E. Topol (Scripps Research Institute), D. Dodwell (Oxford University), M. Cullen (Stanford University) and S. Sammutt (Cambridge University) for their comments.
Author information
Authors and Affiliations
Contributions
A.M.A., D.G., A.L.H., J.R. and M.v.d.S. designed the study. A.M.A. and M.v.d.S. led the development of the automated ML model. A.M.A., D.G., A.L.H. and M.v.d.S. led the writing. D.G., A.L.H., J.R. and M.v.d.S. led the analysis and interpretation of the data. A.M.A. and D.G. provided statistical and analytical support. All authors read and approved the final draft of the manuscript. All authors are accountable for all aspects of the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Machine Intelligence thanks Morteza Noshad and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Alaa, A.M., Gurdasani, D., Harris, A.L. et al. Machine learning to guide the use of adjuvant therapies for breast cancer. Nat Mach Intell 3, 716–726 (2021). https://doi.org/10.1038/s42256-021-00353-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-021-00353-8
This article is cited by
-
Reply to: PREDICT underestimates survival of patients with HER2-positive early-stage breast cancer
npj Breast Cancer (2023)
-
Multiple stakeholders drive diverse interpretability requirements for machine learning in healthcare
Nature Machine Intelligence (2023)
-
Matters Arising: PREDICT underestimates survival of patients with HER2-positive early-stage breast cancer
npj Breast Cancer (2023)
-
Artificial Intelligence in Physical Sciences: Symbolic Regression Trends and Perspectives
Archives of Computational Methods in Engineering (2023)