Abstract
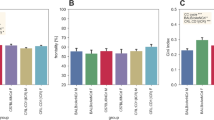
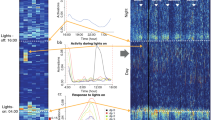
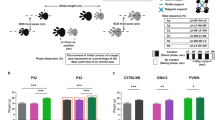
Mouse strains differ markedly in all behaviors, independently of their genetic background. We undertook this study to disentangle the diurnal activity and feature key aspects of three non-genetically altered mouse strains widely used in research, C57BL/6NCrl (inbred), BALB/cAnNCrl (inbred) and CRL:CD1(ICR) (outbred). With this aim, we conducted a longitudinal analysis of the spontaneous locomotor activity of the mice during a 24-h period for 2 months, in two different periods of the year to reduce the seasonality effect. Mice (males and females) were group-housed in Digital Ventilated Cages (Tecniplast), mimicking standard housing conditions in research settings and avoiding the potential bias provided in terms of locomotor activity by single housing. The recorded locomotor activity was analyzed by relying on different and commonly used circadian metrics (i.e., day and night activity, diurnal activity, responses to lights-on and lights-off phases, acrophase and activity onset and regularity disruption index) to capture key behavioral responses for each strain. Our results clearly demonstrate significant differences in the circadian activity of the three selected strains, when comparing inbred versus outbred as well as inbred strains (C57BL/6NCrl versus BALB/cAnNCrl). Conversely, males and females of the same strain displayed similar motor phenotypes; significant differences were recorded only for C57BL/6NCrl and CRL:CD1(ICR) females, which displayed higher average locomotor activity from prepuberty to adulthood. All strain-specific differences were further confirmed by an unsupervised machine learning approach. Altogether, our data corroborate the concept that each strain behaves under characteristic patterns, which needs to be taken into consideration in the study design to ensure experimental reproducibility and comply with essential animal welfare principles.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout










Similar content being viewed by others
Data availability
Datasets and codes used in the analyses are stored at the authors’ home institution and will be provided upon request.
References
Nwagwu, C. D. et al. Endpoint in ovarian cancer xenograft model predicted by nighttime motion metrics. Lab Anim. (NY) 49, 227–232 (2020).
Hillar, C., Onnis, G., Rhea, D. & Tecott, L. Active state organization of spontaneous behavioral patterns. Sci. Rep. 8, 1064 (2018).
Stojakovic, A. et al. Several behavioral traits relevant for alcoholism are controlled by γ2 subunit containing GABAA receptors on dopamine neurons in mice. Neuropsychopharmacology 43, 1548–1556 (2018).
Kamei, J. et al. Effects of diabetes on spontaneous locomotor activity in mice. Neurosci. Lett. 178, 69–72 (1994).
Eckel-Mahan, K. & Sassone-Corsi, P. Phenotyping circadian rhythms in mice. Curr. Protoc. Mouse Biol. 5, 271–281 (2015).
Golini, E. et al. A non-invasive digital biomarker for the detection of rest disturbances in the SOD1G93A mouse model of ALS. Front. Neurosci. 14, 896 (2020).
Loos, M., Verhage, M., Spijker, S. & Smit, A. B. Complex genetics of behavior: BXDs in the automated home-cage. Methods Mol. Biol. 1488, 519–530 (2017).
Hossain, S. M., Wong, B. K. Y. & Simpson, E. M. The dark phase improves genetic discrimination for some high throughput mouse behavioral phenotyping. Genes Brain Behav 3, 167–177 (2004).
Schwartz, W. J. & Zimmerman, P. Circadian timekeeping in BALB/c and C57BL/6 inbred mouse strains. J. Neurosci. 10, 3685–3694 (1990).
Russell, W. M. S. & Burch, R. L. The Principles of Humane Experimental Technique. (Methuen,, London, UK, 1959).
Tam, W. Y. & Cheung, K. K. Phenotypic characteristics of commonly used inbred mouse strains. J. Mol. Med. (Berl.) 98, 1215–1234 (2020).
Bryant C. D. et al. Reduced complexity cross design for behavioral genetics. Molecular-Genetic and Statistical Techniques for Behavioral and Neural Research (ed. Gerlai, R. T.) 165–190 (Elsevier, London, UK, 2018).
Recordati, C. et al. Long-term study on the effects of housing C57BL/6NCrl mice in cages equipped with wireless technology generating extremely low-intensity electromagnetic fields. Toxicol. Pathol. 47, 598–611 (2019).
Burman, O., Marsella, G., Di Clemente, A. & Cervo, L. The effect of exposure to low frequency electromagnetic fields (EMF) as an integral part of the housing system on anxiety-related behaviour, cognition and welfare in two strains of laboratory mouse. PLoS One 13, e0197054 (2018).
Dutta, S. & Sengupta, P. Men and mice: relating their ages. Life Sci. 152, 244–248 (2016).
Gerdin, A.-K. et al. Experimental and husbandry procedures as potential modifiers of the results of phenotyping tests. Physiol. Behav. 106, 602–611 (2012).
Rasmussen, S., Miller, M. M., Filipski, S. B. & Tolwani, R. J. Cage change influences serum corticosterone and anxiety-like behaviors in the mouse. J. Am. Assoc. Lab. Anim. Sci. 50, 479–483 (2011).
Arakawa, H., Blanchard, D. C., Arakawa, K., Dunlap, C. & Blanchard, R. J. Scent marking behavior as an odorant communication in mice. Neurosci. Biobehav. Rev. 32, 1236–1248 (2008).
Iannello, F. Non-intrusive high throughput automated data collection from the home cage. Heliyon 5, e01454 (2019).
Pernold, K. et al. Towards large scale automated cage monitoring—diurnal rhythm and impact of interventions on in-cage activity of C57BL/6J mice recorded 24/7 with a non-disrupting capacitive-based technique. PLoS One 14, e0211063 (2018).
Bains, R. S. et al. Assessing mouse behaviour throughout the light/dark cycle using automated in-cage analysis tools. J. Neurosci. Methods 300, 37–47 (2018).
de Visser, L., van den Bos, R. & Spruijt, B. M. Automated home cage observations as a tool to measure the effects of wheel running on cage floor locomotion. Behav. Brain Res. 160, 382–388 (2005).
Valletta, J. J., Torney, C., Kings, M., Thornton, A. & Madden, J. Applications of machine learning in animal behaviour studies. Anim. Behav. 124, 203–220 (2017).
de Visser, L., van den Bos, R., Kuurman, W. W., Kas, M. J. H. & Spruijt, B. M. Novel approach to the behavioural characterization of inbred mice: automated home cage observations. Genes Brain Behav. 5, 458–466 (2006).
van der Horst, G. T. et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398, 627–630 (1999).
Kopp, C. Locomotor activity rhythm in inbred strains of mice: implications for behavioural studies. Behav. Brain Res. 125, 93–96 (2001).
Simon, M. M. et al. A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol. 14, R82 (2013).
Capri, K. M. et al. Male C57BL6/N and C57BL6/J mice respond differently to constant light and running-wheel access. Front. Behav. Neurosci. 13, 268 (2019).
Sultana, R., Ogundele, O. M. & Lee, C. C. Contrasting characteristic behaviours among common laboratory mouse strains. R. Soc. Open Sci. 6, 190574 (2019).
Kim, D., Chae, S., Lee, J., Yang, H. & Shin, H. S. Variations in the behaviors to novel objects among five inbred strains of mice. Genes Brain Behav. 4, 302–306 (2005).
Sankoorikal, G. M., Kaercher, K. A., Boon, C. J., Lee, J. K. & Brodkin, E. S. A mouse model system for genetic analysis of sociability: C57BL/6J versus BALB/cJ inbred mouse strains. Biol. Psychiatry 59, 415–423 (2006).
Giles, J. M., Whitaker, J. W., Moy, S. S. & Fletcher, C. A. Effect of environmental enrichment on aggression in BALB/cJ and BALB/cByJ mice monitored by using an automated system. J. Am. Assoc. Lab. Anim. Sci. 57, 236–243 (2018).
Aujnarain, A. B., Luo, O. D., Taylor, N., Lai, J. K. Y. & Foster, J. A. Effects of exercise and enrichment on behaviour in CD-1 mice. Behav. Brain Res. 342, 43–50 (2018).
Lightfoot, J. T., Turner, M. J., Daves, M., Vordermark, A. & Kleeberger, S. R. Genetic influence on daily wheel running activity level. Physiol. Genomics 19, 270–276 (2004).
Sherwin, C. M. Voluntary wheel running: a review and novel interpretation. Animal Behav. 56, 11–27 (1998).
Belke, T. W. & McLaughlin, R. J. Habituation contributes to the decline in wheel running within wheel-running reinforcement periods. Behav. Processes 68, 107–115 (2005).
Chia, R., Achilli, F., Festing, M. F. W. & Fisher, E. M. C. The origins and uses of mouse outbred stocks. Nat. Genet. 37, 1181–1186 (2005).
Vannoni, E. et al. Spontaneous behavior in the social homecage discriminates strains, lesions and mutations in mice. J. Neurosci. Methods 234, 26–37 (2014).
Brown, M. J. & Murray, K. A. Phenotyping of genetically engineered mice: humane, ethical, environmental, and husbandry issues. ILAR J. 47, 118–123 (2006).
Tuttle, A. H., Philip, V. M., Chesler, E. J. & Mogil, J. S. Comparing phenotypic variation between inbred and outbred mice. Nat. Methods 15, 994–996 (2018).
Rock, M. L. et al. The time-to-integrate-to-nest test as an indicator of wellbeing in laboratory mice. J. Am. Assoc. Lab. Anim. Sci. 53, 24–28 (2014).
Refinetti, R. Variability of diurnality in laboratory rodents. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 192, 701–714 (2006).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Refinetti, R., Lissen, G. C. & Halberg, F. Procedures for numerical analysis of circadian rhythms. Biol. Rhythm Res. 38, 275–325 (2007).
Diez-Noguera, A. Methods for serial analysis of long time series in the study of biological rhythms. J. Circadian Rhythms 11, 7 (2013).
Brown, L. A., Fisk, A. S., Pothecary, C. A. & Peirson, S. N. Telling the time with a broken clock: quantifying circadian disruption in animal models. Biology (Basel) 8, 18 (2019).
Refinetti, R. Non-parametric procedures for the determination of phase markers of circadian rhythms. Int. J. Biomed. Comput. 30, 49–56 (1992).
Kuznetsova, A., Brockhoff, P. B. & Christensen, R. H. B. lmerTest Package: tests in linear mixed effects models. J. Stat. Softw. 82, 1–26 (2017).
Wolfinger, R. D. Covariance structure selection in general mixed models. Commun. Stat. Simul. Comp 22, 1079–1106 (1993).
Barcikowski, R. S. Statistical power with group mean as the unit of analysis. J. Educ. Behav. Stat. 6, 267–285 (1981).
Acknowledgements
The authors are grateful to Alessandro Grop (CNR-IBBC/EMMA/Infrafrontier/IMPC) and Giampaolo D’Erasmo (CNR-IBBC/EMMA/Infrafrontier/IMPC) for technical assistance with mouse colonies. Further acknowledgements go to (i) the whole management of Charles River Laboratories Italy for the support of Sara Fuochi’s PhD program, providing mice to support the studies described in this paper; (ii) University of Naples Federico II for coordinating the PhD project; and (iii) Tecniplast SpA for providing DVCs.
Author information
Authors and Affiliations
Contributions
S.F., L.D’A., P.d.G. and F.I. designed and supervised the research. M.Ra. and F.S. were responsible for mouse maintenance and prepared a first draft of Methods. M.Ri. and F.I. analyzed and generated the datasets. L.D’A., P.d.G. and S.F. provided a first draft of the manuscript. All the authors reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
F.I. and M.Ri. were employed by Tecniplast SpA, which provided support in the form of salaries for authors F.I. and M.Ri. Tecniplast SpA did not have any additional role in the study design, data collection and analysis, decision to publish or preparation of the manuscript. S.F. was employed by Charles River Laboratories Italy, which provided support in terms of animal models, salary for the author and final review and approval of the manuscript. Charles River Laboratories did not have any additional role in the study design, data collection, analysis or interpretation.
Additional information
Peer review information Lab Animal thanks Rasneer Sonia Bains and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary methods, Supplementary Figures 1–5, activity heatmaps
Supplementary Data 1
Statistical models and test results
Rights and permissions
About this article
Cite this article
Fuochi, S., Rigamonti, M., Iannello, F. et al. Phenotyping spontaneous locomotor activity in inbred and outbred mouse strains by using Digital Ventilated Cages. Lab Anim 50, 215–223 (2021). https://doi.org/10.1038/s41684-021-00793-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41684-021-00793-0