Abstract
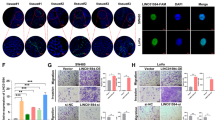
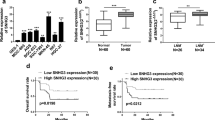
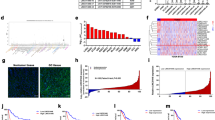
Long non-coding RNAs (lncRNAs) are frequently dysregulated in a variety of human cancers. However, their biological roles in these cancers remain incompletely understood. In this study, we analyze the gene expression profiles of colon cancer tissues and identify a previously unannotated lncRNA, FLJ39051, that we term GSEC (G-quadruplex-forming sequence containing lncRNA), as a lncRNA that is upregulated in colorectal cancer. We further demonstrate that knockdown of GSEC results in the reduction of colon cancer cell motility. We also show that GSEC binds to the DEAH box polypeptide 36 (DHX36) RNA helicase via its G-quadruplex-forming sequence and inhibits DHX36 G-quadruplex unwinding activity. Moreover, knockdown of DHX36 restores the reduced migratory activity of colon cancer cells caused by GSEC knockdown. These results suggest that GSEC plays an important role in colon cancer cell migration by inhibiting the function of DHX36 via its G-quadruplex structure.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N et al. The transcriptional landscape of the mammalian genome. Science 2005; 309: 1559–1563.
Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009; 458: 223–227.
Fatica A, Bozzoni I . Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014; 15: 7–21.
Ulitsky I, Bartel DP . lincRNAs: genomics, evolution, and mechanisms. Cell 2013; 154: 26–46.
Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell 2014; 26: 344–357.
Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K et al. LincRNA-p21 activates p21 in cis to promote polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell 2014; 54: 777–790.
Lam MTY, Li W, Rosenfeld MG, Glass CK . Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci 2014; 39: 170–182.
O’Leary VB, Ovsepian SV, Carrascosa LG, Buske FA, Radulovic V, Niyazi M et al. PARTICLE, a triplex-forming long ncRNA, regulates locus-specific methylation in response to low-dose irradiation. Cell Rep 2015; 11: 474–485.
Arab K, Park YJ, Lindroth AM, Schäfer A, Oakes C, Weichenhan D et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell 2014; 55: 604–614.
Gumireddy K, Li A, Yan J, Setoyama T, Johannes GJ, Orom UA et al. Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. EMBO J 2013; 32: 2672–2684.
Gong C, Maquat LE . lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature 2011; 470: 284–288.
Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 2014; 344: 310–313.
Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP . Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal 2010; 3: ra8.
Liu X, Li D, Zhang W, Guo M, Zhan Q . Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. EMBO J 2012; 31: 4415–4427.
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014; 25: 666–681.
Xing Z, Lin A, Li C, Liang K, Wang S, Liu Y et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 2014; 159: 1110–1125.
Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2012; 493: 231–235.
Lin N, Chang KY, Li Z, Gates K, Rana Z, Dang J et al. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell 2014; 53: 1005–1019.
Bierhoff H, Dammert MA, Brocks D, Dambacher S, Schotta G, Grummt I . Quiescence-induced lncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol Cell 2014; 54: 675–682.
Richards EJ, Zhang G, Li Z-P, Permuth-Wey J, Challa S, Li Y et al. Long non-coding RNAs (LncRNA) regulated by transforming growth factor (TGF) β. J Biol Chem 2015; 290: 6857–6867.
Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature 2013; 500: 598–602.
Takayama K-I, Horie-Inoue K, Katayama S, Suzuki T, Tsutsumi S, Ikeda K et al. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J 2013; 32: 1665–1680.
Xiang J-F, Yin Q-F, Chen T, Zhang Y, Zhang X-O, Wu Z et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res 2014; 24: 513–531.
Graham LD, Pedersen SK, Brown GS, Ho T, Kassir Z, Moynihan AT et al. Colorectal neoplasia differentially expressed (CRNDE), a novel gene with elevated expression in colorectal adenomas and adenocarcinomas. Genes Cancer 2011; 2: 829–840.
Prensner JR, Iyer MK, Sahu A, Asangani Ia, Cao Q, Patel L et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet 2013; 45: 1392–1398.
Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell 2013; 49: 1083–1096.
Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu M et al. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis 2014; 5: e1008.
Sun M, Gadad SS, Kim D-S, Kraus WL . Discovery, annotation, and functional analysis of long noncoding RNAs controlling cell-cycle gene expression and proliferation in breast cancer cells. Mol Cell 2015; 59: 1–14.
Gutschner T, Diederichs S . The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol 2012; 9: 703–719.
Tseng Y-Y, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H et al. PVT1 dependence in cancer with MYC copy-number increase. Nature 2014; 512: 82.
Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S . Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res 2006; 34: 5402–5415.
Millevoi S, Moine H, Vagner S . G-quadruplexes in RNA biology. Wiley Interdiscip Rev RNA 2012; 3: 495–507.
Huang W, Smaldino PJ, Zhang Q, Miller LD, Cao P, Stadelman K et al. Yin Yang 1 contains G-quadruplex structures in its promoter and 5′-UTR and its expression is modulated by G4 resolvase 1. Nucleic Acids Res 2012; 40: 1033–1049.
Jayaraj GG, Pandey S, Scaria V, Maiti S . Potential G-quadruplexes in the human long non-coding transcriptome. RNA Biol 2012; 9: 81–89.
Tsang WP, Ng EKO, Ng SSM, Jin H, Yu J, Sung JJY et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis 2010; 31: 350–358.
Vaughn JP, Creacy SD, Routh ED, Joyner-Butt C, Jenkins GS, Pauli S et al. The DEXH protein product of the DHX36 gene is the major source of tetramolecular quadruplex G4-DNA resolving activity in HeLa cell lysates. J Biol Chem 2005; 280: 38117–38120.
Booy EP, Howard R, Marushchak O, Ariyo EO, Meier M, Novakowski SK et al. The RNA helicase RHAU (DHX36) suppresses expression of the transcription factor PITX1. Nucleic Acids Res 2014; 42: 3346–3361.
Kim H-N, Lee J-H, Bae S-C, Ryoo H-M, Kim H-H, Ha H et al. Histone deacetylase inhibitor MS-275 stimulates bone formation in part by enhancing Dhx36-mediated TNAP transcription. J Bone Miner Res 2011; 26: 2161–2173.
Booy EP, Meier M, Okun N, Novakowski SK, Xiong S, Stetefeld J et al. The RNA helicase RHAU (DHX36) unwinds a G4-quadruplex in human telomerase RNA and promotes the formation of the P1 helix template boundary. Nucleic Acids Res 2012; 40: 4110–4124.
Höck J, Weinmann L, Ender C, Rüdel S, Kremmer E, Raabe M et al. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep 2007; 8: 1052–1060.
Bicker S, Khudayberdiev S, Weiß K, Zocher K, Baumeister S, Schratt G . The DEAH-box helicase DHX36 mediates dendritic localization of the neuronal precursor-microRNA-134. Genes Dev 2013; 27: 991–996.
Kawasaki Y, Sato R, Akiyama T . Mutated APC and Asef are involved in the migration of colorectal tumour cells. Nat Cell Biol 2003; 5: 211–215.
Creacy SD, Routh ED, Iwamoto F, Nagamine Y, Akman SA, Vaughn JP . G4 resolvase 1 binds both DNA and RNA tetramolecular quadruplex with high affinity and is the major source of tetramolecular quadruplex G4-DNA and G4-RNA resolving activity in HeLa cell lysates. J Biol Chem 2008; 283: 34626–34634.
Yanagida S, Taniue K, Sugimasa H, Nasu E, Takeda Y, Kobayashi M et al. ASBEL, an ANA/BTG3 antisense transcript required for tumorigenicity of ovarian carcinoma. Sci Rep 2013; 3: 1305.
Acknowledgements
This work was supported by Grants-in-Aid for Scientific Research on Innovative Areas (Integrative Research on Cancer Microenvironment Network) and Research Program of the Project for Development of Innovative Research on Cancer Therapeutics (P-Direct), Ministry of Education, Culture, Sports, Science and Technology of Japan and Japan Society for the Promotion of Science Grant-in-Aid for JSPS Fellows (Grant Number 13J08104).
Author Contributions
KM, MM, YK, JN and SS performed the experiments. LN performed mass spectrometry analysis. KM, YK and TA analyzed the data and wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Matsumura, K., Kawasaki, Y., Miyamoto, M. et al. The novel G-quadruplex-containing long non-coding RNA GSEC antagonizes DHX36 and modulates colon cancer cell migration. Oncogene 36, 1191–1199 (2017). https://doi.org/10.1038/onc.2016.282
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.282
This article is cited by
-
Identification and Validation of Basement Membrane Related LncRNA Signatures as a Novel Prognostic Model for Hepatocellular Carcinoma
Biochemical Genetics (2024)
-
Long non-coding RNA signature in colorectal cancer: research progression and clinical application
Cancer Cell International (2023)
-
G-quadruplexes from non-coding RNAs
Journal of Molecular Medicine (2023)
-
Interaction between non-coding RNAs, mRNAs and G-quadruplexes
Cancer Cell International (2022)
-
Long noncoding RNA GSEC promotes neutrophil inflammatory activation by supporting PFKFB3-involved glycolytic metabolism in sepsis
Cell Death & Disease (2021)