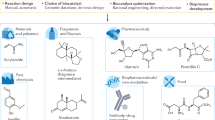
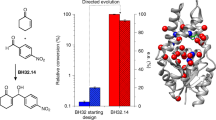
Abstract
Over the past ten years, scientific and technological advances have established biocatalysis as a practical and environmentally friendly alternative to traditional metallo- and organocatalysis in chemical synthesis, both in the laboratory and on an industrial scale. Key advances in DNA sequencing and gene synthesis are at the base of tremendous progress in tailoring biocatalysts by protein engineering and design, and the ability to reorganize enzymes into new biosynthetic pathways. To highlight these achievements, here we discuss applications of protein-engineered biocatalysts ranging from commodity chemicals to advanced pharmaceutical intermediates that use enzyme catalysis as a key step.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Buchholz, K., Kasche, V. & Bornscheuer, U. T. Biocatalysts and Enzyme Technology 2nd edn (Wiley-VCH, 2012)
Drauz, K., Gröger, H., May, O. (eds) Enzyme Catalysis in Organic Synthesis Vols 1–3, 3rd edn (Wiley-VCH, 2012)
Bornscheuer, U. T. & Kazlauskas, R. J. Hydrolases in Organic Synthesis - Regio- and Stereoselective Biotransformations 2nd edn (Wiley-VCH, 2006)
Liese, A., Seelbach, K., Wandrey, C. (eds) Industrial Biotransformations 2nd edn (Wiley-VCH, 2006)
Wenda, S., Illner, S., Mell, A. & Kragl, U. Industrial biotechnology—the future of green chemistry? Green Chem. 13, 3007–3047 (2011)
Rosenthaler, L. Durch Enzyme bewirkte asymmetrische Synthese. Biochem. Z. 14, 238–253 (1908)
Sedlaczek, L. Biotransformations of steroids. Crit. Rev. Biotechnol. 7, 187–236 (1988)
Estell, D. A., Graycar, T. P. & Wells, J. A. Engineering an enzyme by site-directed mutagenesis to be resistant to chemical oxidation. J. Biol. Chem. 260, 6518–6521 (1985)This paper puts forward the basis for the first application of protein engineering in industrial biotechnology.
Jensen, V. J. & Rugh, S. Industrial scale production and application of immobilized glucose isomerase. Methods Enzymol. 136, 356–370 (1987)
Bruggink, A., Roos, E. C. & de Vroom, E. Penicillin acylase in the industrial production of β-lactam antibiotics. Org. Process Res. Dev. 2, 128–133 (1998)
Griengl, H., Schwab, H. & Fechter, M. The synthesis of chiral cyanohydrins by oxynitrilases. Trends Biotechnol. 18, 252–256 (2000)
Hills, G. Industrial use of lipases to produce fatty acid esters. Eur. J. Lipid Sci. Technol. 105, 601–607 (2003)
Nagasawa, T., Nakamura, T. & Yamada, H. Production of acrylic acid and methacrylic acid using Rhodococcus rhodochrous J1 nitrilase. Appl. Microbiol. Biotechnol. 34, 322–324 (1990)
Francis, J. C. & Hansche, P. E. Directed evolution of metabolic pathways in microbial populations. I. Modification of the acid phosphatase pH optimum in S. cerevisiae.. Genetics 70, 59–73 (1972)
Nakamura, C. E. & Whited, G. M. Metabolic engineering for the microbial production of 1,3-propanediol. Curr. Opin. Biotechnol. 14, 454–459 (2003)
Liang, J. et al. Development of a biocatalytic process as an alternative to the (-)-DIP-Cl-mediated asymmetric reduction of a key intermediate of montelukast. Org. Process Res. Dev. 14, 193–198 (2010)
Savile, C. K. et al. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science 329, 305–309 (2010)
Desai, A. A. Sitagliptin manufacture: a compelling tale of green chemistry, process intensification, and industrial asymmetric catalysis. Angew. Chem. Int. Ed. 50, 1974–1976 (2011)This highlight article compares in detail two processes for the production of sitagliptin, one catalysed by rhodium and one catalysed by a transaminase.
O’Maille, P. E. et al. Quantitative exploration of the catalytic landscape separating divergent plant sesquiterpene synthases. Nature Chem. Biol. 4, 617–623 (2008)
Atsumi, S., Hanai, T. & Liao, J. C. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451, 86–89 (2008)This paper describes the efficient diversion of amino-acid metabolism to the production of alcohols.
Lee, S. K., Chou, H., Ham, T. S., Lee, T. S. & Keasling, J. D. Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr. Opin. Biotechnol. 19, 556–563 (2008)
Steen, E. J. et al. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463, 559–562 (2010)
Bohmert-Tatarev, K., McAvoy, S., Daughtry, S., Peoples, O. P. & Snell, K. D. High levels of bioplastic are produced in fertile transplastomic tobacco plants engineered with a synthetic operon for the production of polyhydroxybutyrate. Plant Physiol. 155, 1690–1708 (2011)
McKenna, R. & Nielsen, D. R. Styrene biosynthesis from glucose by engineered E. coli. Metab. Eng. 13, 544–554 (2011)
Kazlauskas, R. J. & Bornscheuer, U. T. Finding better protein engineering strategies. Nature Chem. Biol. 5, 526–529 (2009)
Lutz, S., Bornscheuer, U. T. (eds) Protein Engineering Handbook (Wiley-VCH, 2009)
Turner, N. J. Directed evolution drives the next generation of biocatalysts. Nature Chem. Biol. 5, 567–573 (2009)
Röthlisberger, D. et al. Kemp elimination catalysts by computational enzyme design. Nature 453, 190–195 (2008)
Schmid, A. et al. Industrial biocatalysis today and tomorrow. Nature 409, 258–268 (2001)
Arnold, F. H. Combinatorial and computational challenges for biocatalyst design. Nature 409, 253–257 (2001)
Schoemaker, H. E., Mink, D. & Wubbolts, M. G. Dispelling the myths—biocatalysis in industrial synthesis. Science 299, 1694–1697 (2003)
Keasling, J. D. Manufacturing molecules through metabolic engineering. Science 330, 1355–1358 (2010)
Madison, L. L. & Huisman, G. W. Metabolic engineering of poly(3-hydroxy-alkanoates): from DNA to plastic. Microbiol. Mol. Biol. Rev. 63, 21–53 (1999)
Wetterstrand, K. A. DNA sequencing costs: data from the NHGRI large-scale genome sequencing program. National Human Genome Research Project 〈http://www.genome.gov/sequencingcosts〉 (2011)
Lorenz, P. & Eck, J. Metagenomics and industrial applications. Nature Rev. Microbiol. 3, 510–516 (2005)
Fowler, D. M. et al. High-resolution mapping of protein sequence- function relationships. Nature Methods 7, 741–746 (2010)
Richmond, K. E. et al. Amplification and assembly of chip-eluted DNA (AACED): a method for high-throughput gene synthesis. Nucleic Acids Res. 32, 5011–5018 (2004)
Gibson, D. G. et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329, 52–56 (2010)
Lutz, S. Beyond directed evolution—semi-rational protein engineering and design. Curr. Opin. Biotechnol. 21, 734–743 (2010)
Höhne, M., Schätzle, S., Jochens, H., Robins, K. & Bornscheuer, U. T. Rational assignment of key motifs for function guides in silico enzyme identification. Nature Chem. Biol. 6, 807–813 (2010)In this paper, careful analysis of key motifs of 5,000 pyridoxal-5-phosphate-dependent transaminase sequences in public databases identified 20 novel enzymes for which substrate preference (ketone, not α-keto acid) and enantiopreference (( R ), not ( S )) could be predicted and experimentally confirmed.
Jochens, H. & Bornscheuer, U. T. Natural diversity to guide focused directed evolution. ChemBioChem 11, 1861–1866 (2010)
Park, S. et al. Focusing mutations into the P. fluorescens esterase binding site increases enantioselectivity more effectively than distant mutations. Chem. Biol. 12, 45–54 (2005)
Horsman, G. P., Liu, A. M. F., Henke, E., Bornscheuer, U. T. & Kazlauskas, R. J. Mutations in distant residues moderately increase the enantioselectivity of Pseudomonas fluorescens esterase towards methyl 3-bromo-2-methylpropanoate and ethyl 3-phenylbutyrate. Chemistry 9, 1933–1939 (2003)
Esvelt, K. M., Carlson, J. C. & Liu, D. R. A system for the continuous directed evolution of biomolecules. Nature 472, 499–503 (2011)
Moore, J. C., Pollard, D. J., Kosjek, B. & Devine, P. N. Advances in the enzymatic reduction of ketones. Acc. Chem. Res. 40, 1412–1419 (2007)
Strohmeier, G. A., Pichler, H., May, O. & Gruber-Khadjawi, M. Application of designed enzymes in organic synthesis. Chem. Rev. 111, 4141–4164 (2011)This is a review about protein engineering to create biocatalysts for industrial applications.
Wiese, A., Wilms, B., Syldatk, C., Mattes, R. & Altenbuchner, J. Cloning, nucleotide sequence and expression of a hydantoinase and carbamoylase gene from Arthrobacter aurescens DSM 3745 in Escherichia coli and comparison with the corresponding genes from Arthrobacter aurescens DSM 3747. Appl. Microbiol. Biotechnol. 55, 750–757 (2001)
Martinez-Gomez, A. I. et al. Recombinant polycistronic structure of hydantoinase process genes in Escherichia coli for the production of optically pure D-amino acids. Appl. Environ. Microbiol. 73, 1525–1531 (2007)
Panke, S. & Wubbolts, M. Advances in biocatalytic synthesis of pharmaceutical intermediates. Curr. Opin. Chem. Biol. 9, 188–194 (2005)
Breuer, M. et al. Industrial methods for the production of optically active intermediates. Angew. Chem. Int. Ed. 43, 788–824 (2004)
DeSantis, G. et al. Creation of a productive, highly enantioselective nitrilase through gene site saturation mutagenesis (GSSM). J. Am. Chem. Soc. 125, 11476–11477 (2003)This paper describes how a single amino-acid substitution can create a nitrilase with high enantioselectivity at 3 M substrate concentration, for synthesis of an intermediate for atorvastatin.
Ma, S. K. et al. A green-by-design biocatalytic process for atorvastatin intermediate. Green Chem. 12, 81–86 (2010)
Anastas, P., Warner, J. (eds) Green Chemistry: Theory and Practice (Oxford Univ. Press, 1998)
Yang, T. H. et al. Biosynthesis of polylactic acid and its copolymers using evolved propionate CoA transferase and PHA synthase. Biotechnol. Bioeng. 105, 150–160 (2010)
Bernath, K. et al. In vitro compartmentalization by double emulsions: sorting and gene enrichment by fluorescence activated cell sorting. Anal. Biochem. 325, 151–157 (2004)
Becker, S. et al. Single-cell high-throughput screening to identify enantioselective hydrolytic enzymes. Angew. Chem. Int. Ed. 47, 5085–5088 (2008)
Fernández-Alvaro, E. et al. A combination of in vivo selection and cell sorting for the identification of enantioselective biocatalysts. Angew. Chem. Int. Ed. 50, 8584–8587 (2011)
Whittle, E. & Shanklin, J. Engineering delta 9–16:0-acyl carrier protein (ACP) desaturase specificity based on combinatorial saturation mutagenesis and logical redesign of the castor delta 9–18:0-ACP desaturase. J. Biol. Chem. 276, 21500–21505 (2001)
Seelig, B. & Szostak, J. W. Selection and evolution of enzymes from a partially randomized non-catalytic scaffold. Nature 448, 828–831 (2007)
Fox, R. J. et al. Improving catalytic function by ProSAR-driven enzyme evolution. Nature Biotechnol. 25, 338–344 (2007)
Yoshikuni, Y., Ferrin, T. E. & Keasling, J. D. Designed divergent evolution of enzyme function. Nature 440, 1078–1082 (2006)
Bava, K. A., Gromiha, M. M., Uedaira, H. & Kitajima, K. &. Sarai, A. ProTherm, version 4.0: thermodynamic database for proteins and mutants. Nucleic Acids Res. 32, D120–D121 (2004)
Guo, H. H., Choe, J. & Loeb, L. A. Protein tolerance to random amino acid change. Proc. Natl Acad. Sci. USA 101, 9205–9210 (2004)This paper shows that about 34% of random amino-acid replacements inactivate a protein’s functions, indicating the importance of starting with a stabilized protein for protein engineering experiments.
Drummond, D. A., Bloom, J. D., Adami, C., Wilke, C. O. & Arnold, F. H. Why highly expressed proteins evolve slowly. Proc. Natl Acad. Sci. USA 102, 14338–14343 (2005)
Gupta, R. D. & Tawfik, D. S. Directed enzyme evolution via small and effective neutral drift libraries. Nature Methods 5, 939–942 (2008)
Bloom, J. D., Romero, P. A., Lu, Z. & Arnold, F. H. Neutral genetic drift can alter promiscuous protein functions, potentially aiding functional evolution. Biol. Direct 2, 17 (2007)
Wells, J. A. Additivity of mutational effects in proteins. Biochemistry 29, 8509–8517 (1990)
Moore, J. C. & Arnold, F. H. Directed evolution of a para-nitrobenzyl esterase for aqueous-organic solvents. Nature Biotechnol. 14, 458–467 (1996)
Reetz, M. T., Zonta, A., Schimossek, K., Liebeton, K. & Jaeger, K.-E. Creation of enantioselective biocatalysts for organic chemistry by in vitro evolution. Angew. Chem. Int. Edn Engl. 36, 2830–2832 (1997)
Weinreich, D. M., Delaney, N. F., Depristo, M. A. & Hartl, D. L. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312, 111–114 (2006)
Reetz, M. T. & Sanchis, J. Constructing and analyzing the fitness landscape of an experimental evolutionary process. ChemBioChem 9, 2260–2267 (2008)
Jiang, L. et al. De novo computational design of retro-aldol enzymes. Science 319, 1387–1391 (2008)
Meyer, D. et al. Conversion of pyruvate decarboxylase into an enantioselective carboligase with biosynthetic potential. J. Am. Chem. Soc. 133, 3609–3616 (2011)
Siegel, J. B. et al. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction. Science 329, 309–313 (2010)
Randolph, J., Yagodkin, A., Lamaitre, M., Azhayev, A. & Mackie, H. Codon based mutagenesis using trimer phosphoramidites. Nucleic Acids Symp. Ser. 52, 479 (2008)
Rana, S., Yeh, Y. C. & Rotello, V. M. Engineering the nanoparticle-protein interface: applications and possibilities. Curr. Opin. Chem. Biol. 14, 828–834 (2010)
Dueber, J. E. et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nature Biotechnol. 27, 753–759 (2009)
McDonald, T. J. et al. Wiring-up hydrogenase with single-walled carbon nanotubes. Nano Lett. 7, 3528–3534 (2007)
Mouse Genome Sequencing Consortium Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002)
Savile, C. K. & Lalonde, J. J. Biotechnology for the acceleration of carbon dioxide capture and sequestration. Curr. Opin. Biotechnol. 22, 818–823 (2011)
Morikawa, S. et al. Highly active mutants of carbonyl reductase S1 with inverted coenzyme specificity and production of optically active alcohols. Biosci. Biotechnol. Biochem. 69, 544–552 (2005)
Patel, R. N., Chu, L. & Mueller, R. Diastereoselective microbial reduction of (S)-[3-chloro-2-oxo-1-(phenylmethyl)propyl]carbamic acid, 1,1-dimethylethyl ester. Tetrahedr. Asymm. 14, 3105–3109 (2003)
Liang, J. et al. Highly enantioselective reduction of a small heterocyclic ketone: biocatalytic reduction of tetrahydrothiophene-3-one to the corresponding (R)-alcohol. Org. Process Res. Dev. 14, 188–192 (2010)
Urano, N. et al. Directed evolution of an aminoalcohol dehydrogenase for efficient production of double chiral aminoalcohols. J. Biosci. Bioeng. 111, 266–271 (2011)
Gooding, O. W. et al. Development of a practical biocatalytic process for (R)-2-methylpentanol. Org. Process Res. Dev. 14, 119–126 (2010)
Cobley, C. J., Hanson, C. H., Lloyd, M. C. & Simmonds, S. The combination of hydroformylation and biocatalysis for the large-scale synthesis of (S)-allysine ethylene acetal. Org. Process Res. Dev. 15, 284–290 (2011)
Xie, X., Watanabe, K., Wojcicki, W. A., Wang, C. C. & Tang, Y. Biosynthesis of lovastatin analogs with a broadly specific acyltransferase. Chem. Biol. 13, 1161–1169 (2006)
Truppo, M. D. & Hughes, G. Development of an improved immobilized CAL-B for the enzymatic resolution of a key intermediate to odanacatib. Org. Process Res. Dev. 15, 1033–1035 (2011)
Brocklehurst, C. E., Laumen, K., Vecchia, L. L., Shaw, D. & Vögtle, M. Diastereoisomeric salt formation and enzyme-catalyzed kinetic resolution as complementary methods for the chiral separation of cis-/trans-enantiomers of 3-aminocyclohexanol. Org. Process Res. Dev. 15, 294–300 (2011)
Iding, H. et al. in Asymmetric Catalysis on Industrial Scale (eds Blaser, H. U. & Federsel, H. J. ) 377–396 (Wiley-VCH, 2010)
Chen, Y. J. et al. Enzymatic preparation of an (S)-amino acid from a racemic amino acid. Org. Process Res. Dev. 15, 241–248 (2011)
Rousseau, A. L. et al. Scale-up of a chemo-biocatalytic route to (2R,4R)- and (2S,4S)-monatin. Org. Process Res. Dev. 15, 249–257 (2011)
Greenberg, W. A. et al. Development of an efficient, scalable, aldolase-catalyzed process for enantioselective synthesis of statin intermediates. Proc. Natl Acad. Sci. USA 101, 5788–5793 (2004)
Horinouchi, N. et al. Biochemical retrosynthesis of 2′-deoxyribonucleosides from glucose, acetaldehyde, and a nucleobase. Appl. Microbiol. Biotechnol. 71, 615–621 (2006)
Hibi, M. et al. Characterization of Bacillus thuringiensis L-isoleucine dioxygenase for production of useful amino acids. Appl. Environ. Microbiol. 77, 6926–6930 (2011)
de Lange, B. et al. Asymmetric synthesis of (S)-2-indoline carboxylic acid by combining biocatalysis and homogeneous catalysis. ChemCatChem 3, 289–292 (2011)
Asano, Y., Mihara, Y. & Yamada, H. A new enzymatic method of selective phosphorylation of nucleosides. J. Mol. Catal. B 6, 271–277 (1999)
Hermann, M. et al. Alternative pig liver esterase (APLE) - cloning, identification and functional expression in Pichia pastoris of a versatile new biocatalyst. J. Biotechnol. 133, 301–310 (2008)
Ikenaka, Y. et al. Thermostability reinforcement through a combination of thermostability-related mutations of N-carbamyl-D-amino acid amidohydrolase. Biosci. Biotechnol. Biochem. 63, 91–95 (1999)
Ro, D. K. et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440, 940–943 (2006)
Acknowledgements
We thank H.-P. Meyer and R. Fox for discussions. R.J.K. thanks the US National Science Foundation (CBET-0932762) and the Korea Science and Engineering Foundation funded by the Ministry of Education, Science and Technology (WCU programme R32-2008-000-10213-0). U.T.B. and S.L. thank the German Research Foundation (SPP 1170, Bo1864/4-1) and, respectively, the US National Science Foundation (CBET-0730312) for financial support.
Author information
Authors and Affiliations
Contributions
U.T.B., R.J.K. and S.L. drafted the text; K.R., G.W.H. and J.C.M. collected the examples for industrial applications; and the authors together wrote and edited the review. All authors contributed equally.
Corresponding author
Ethics declarations
Competing interests
K.R. is an employee of Lonza AG, G.W.H. is an employee of Codexis Inc. and J.C.M. is an employee of Merck & Co. Inc. All three companies are working on biocatalysis.
Rights and permissions
About this article
Cite this article
Bornscheuer, U., Huisman, G., Kazlauskas, R. et al. Engineering the third wave of biocatalysis. Nature 485, 185–194 (2012). https://doi.org/10.1038/nature11117
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11117
This article is cited by
-
Biocatalytic strategy for the construction of sp3-rich polycyclic compounds from directed evolution and computational modelling
Nature Chemistry (2024)
-
Effective engineering of a ketoreductase for the biocatalytic synthesis of an ipatasertib precursor
Communications Chemistry (2024)
-
Remote stereocontrol with azaarenes via enzymatic hydrogen atom transfer
Nature Chemistry (2024)
-
Ligninolytic and cellulolytic enzymes — biocatalysts for green agenda
Biomass Conversion and Biorefinery (2024)
-
Deep Insight of Design, Mechanism, and Cancer Theranostic Strategy of Nanozymes
Nano-Micro Letters (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.