Abstract
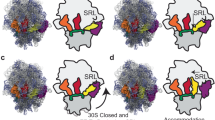
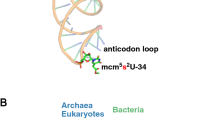
The natural modification of specific nucleosides in many tRNAs is essential during decoding of mRNA by the ribosome. For example, tRNALysUUU requires the modification N6-threonylcarbamoyladenosine at position 37 (t6A37), adjacent and 3′ to the anticodon, to bind AAA in the A site of the ribosomal 30S subunit. Moreover, it can only bind both AAA and AAG lysine codons when doubly modified with t6A37 and either 5-methylaminomethyluridine or 2-thiouridine at the wobble position (mnm5U34 or s2U34). Here we report crystal structures of modified tRNA anticodon stem-loops bound to the 30S ribosomal subunit with lysine codons in the A site. These structures allow the rationalization of how modifications in the anticodon loop enable decoding of both lysine codons AAA and AAG.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ogle, J.M., Carter, A.P. & Ramakrishnan, V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci. 28, 259–266 (2003).
Ogle, J.M. et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902 (2001).
Ogle, J.M., Murphy, F.V., Tarry, M.J. & Ramakrishnan, V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111, 721–732 (2002).
Crick, F.H.C. Codon-anticodon pairing: the wobble hypothesis. J. Mol. Biol. 19, 548–555 (1966).
Yarian, C. et al. Modified nucleoside dependent Watson-Crick and wobble codon binding by tRNALysUUU species. Biochemistry 39, 13390–13395 (2000).
Agris, P.F. Decoding the genome: a modified view. Nucleic Acids Res. 32, 223–238 (2004).
Sprinzl, M., Horn, C., Brown, M., Loudovitch, A. & Steinberg, S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26, 148–153 (1998).
Krüger, M.K., Pedersen, S., Hagervall, T.G. & Sørensen, M.A. The modification of the wobble base of tRNAGlu modulates the translation rate of glutamic acid codons in vivo. J. Mol. Biol. 284, 621–631 (1998).
Yarian, C. et al. Accurate translation of the genetic code depends on tRNA modified nucleosides. J. Biol. Chem. 277, 16391–16395 (2002).
Urbonavicius, J., Qian, Q., Durand, J.M., Hagervall, T.G. & Björk, G.R. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 20, 4863–4873 (2001).
Phelps, S.S., Malkiewicz, A., Agris, P.F. & Joseph, S. Modified nucleotides in tRNALys and tRNAVal are important for translocation. J. Mol. Biol. 338, 439–444 (2004).
Björk, G.R. In tRNA: Structure, Biosynthesis and Function (eds. Söll, D. & RajBhandary, U.) 165–205 (American Society for Microbiology, Washington, DC, 1995).
von Ahsen, U., Green, R., Schroeder, R. & Noller, H.F. Identification of 2′-hydroxyl groups required for interaction of a tRNA anticodon stem-loop region with the ribosome. RNA 3, 49–56 (1997).
Phelps, S.S., Jerinic, O. & Joseph, S. Universally conserved interactions between the ribosome and the anticodon stem-loop of A site tRNA important for translocation. Mol. Cell 10, 799–807 (2002).
Parthasarathy, R., Ohrt, J.M. & Chheda, G.B. Modified nucleosides and conformation of anticodon loops: crystal structure of t6A and g6A. Biochemistry 16, 4999–5008 (1977).
Stuart, J.W. et al. Functional anticodon architecture of human tRNALys3 includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A. Biochemistry 39, 13396–13404 (2000).
Bénas, P. et al. The crystal structure of HIV reverse-transcription primer tRNA(Lys,3) shows a canonical anticodon loop. RNA 6, 1347–1355 (2000).
Grosjean, H., Söll, D. & Crothers, D.M. Studies of the complex between transfer RNAs with complementary anticodons. J. Mol. Biol. 103, 499–519 (1976).
Konevega, A.L. et al. Purine bases at position 37 of tRNA stabilize codon-anticodon interaction in the ribosomal A site by stacking and Mg2+-dependent interactions. RNA 10, 90–101 (2004).
Chou, S.-H. & Tseng, Y.-Y. Cross-strand purine-pyrimidine stack and sheared purine·pyrimidine pairing in the human HIV-1 reverse transcriptase inhibitors. J. Mol. Biol. 285, 41–48 (1999).
Serra, M.J. & Turner, D.H. Predicting thermodynamic properties of RNA. Methods Enzymol. 259, 242–261 (1995).
Saenger, W. Principles of Nucleic Acid Structure (Springer, New York, 1984).
Battle, D.J. & Doudna, J.A. Specificity of RNA-RNA helix recognition. Proc. Natl. Acad. Sci. USA 99, 11676–11681 (2002).
Agris, P.F. Wobble position modified nucleosides evolved to select transfer RNA codon recognition: a modified-wobble hypothesis. Biochimie 73, 1345–1349 (1991).
Freier, S.M. et al. Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl. Acad. Sci. USA 83, 9373–9377 (1986).
Sundaram, M., Durant, P.C. & Davis, D.R. Hypermodified nucleosides in the anticodon of tRNALys stabilize a canonical U-turn structure. Biochemistry 39, 12575–12584 (2000).
Yokoyama, S. & Nishimura, S. In tRNA: Structure, Biosynthesis, and Function (eds. Söll, D. & RajBhandary, U.) 207–223 (American Society for Microbiology, Washington, DC, 1995).
Takai, K. & Yokoyama, S. Roles of 5-substituents of tRNA wobble uridines in the recognition of purine-ending codons. Nucleic Acids Res. 31, 6383–6391 (2003).
Auffinger, P. & Westhof, E. Singly and bifurcated hydrogen-bonded base-pairs in tRNA anticodon hairpins and ribozymes. J. Mol. Biol. 292, 467–483 (1999).
Yang, J. & Gellman, S.H. Energetic superiority of two-center hydrogen bonding relative to three-center hydrogen bonding in a model system. J. Am. Chem. Soc. 120, 9090–9091 (1998).
Mizuno, H. & Sundaralingam, M. Stacking of Crick wobble pair and Watson-Crick pair: stability rules of G-U pairs at ends of helical stems in tRNAs and the relation to codon-anticodon wobble interaction. Nucleic Acids Res. 5, 4451–4461 (1978).
Chen, X., McDowell, J.A., Kierzek, R., Krugh, T.R. & Turner, D.H. Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G·U pairs: one hydrogen bond for each G·U pair in r(GGCGUGCC)2 and two for each G·U pair in r(GAGUGCUC)2. Biochemistry 39, 8970–8982 (2000).
Rodnina, M.V. & Wintermeyer, W. Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu. Rev. Biochem. 70, 415–435 (2001).
Clemons, W.M. Jr. et al. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: purification, crystallization and structure determination. J. Mol. Biol. 310, 827–843 (2001).
Ogilvie, K.K., Usman, N., Nicoghosian, K. & Cedergren, R.J. Total chemical synthesis of a 77-nucleotide-long RNA sequence having methionine-acceptance activity. Proc. Natl. Acad. Sci. USA 85, 5764–5768 (1988).
Agris, P.F. et al. Site-selected introduction of modified purine and pyrimidine ribonucleosides into RNA by automated phosphoramidite chemistry. Biochimie 77, 125–134 (1995).
Faulstich, K., Wörner, K., Brill, H. & Engels, J.W. A sequencing method for RNA oligonucleotides based on mass spectrometry. Anal. Chem. 69, 4349–4353 (1997).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276A, 307–326 (1997).
Ravelli, R.B.G., Sweet, R.M., Skinner, J.M., Duisenberg, A.J.M. & Kroon, J. STRATEGY: a program to optimize the starting spindle angle and scan range for X-ray data collection. J. Appl. Cryst. 30, 551–554 (1997).
Brünger, A.T. et al. Crystallography & NMR System: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Kleywegt, G.J. & Jones, T.A. Databases in protein crystallography. Acta Crystallogr. D 54, 1119–1131 (1998).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Koradi, R., Billeter, M. & Wüthrich, K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55 (1996).
Schneider, T.R. A genetic algorithm for the identification of conformationally invariant regions in protein molecules. Acta Crystallogr. D 58, 195–208 (2002).
Lu, X.J. & Olson, W.K. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 31, 5108–5121 (2003).
Acknowledgements
The authors acknowledge the contributions of W. Czestkowski for his modified nucleoside synthetic work, W. Graham for his purification of the doubly modified ASL, M. DeRider for contributions to structural modeling and J. Ogle for comments on the manuscript. This work was funded by the Medical Research Council, UK, US National Institutes of Health (NIH) grant GM67624 and a grant from the Agouron Institute (V.R.); US National Science Foundation grant MCB9986011 and NIH grant GM23037 (P.F.A.); and by the grant KBN 7TO9A01721 (A.M.). F.V.M. was supported by a European Molecular Biology Organization long term fellowship.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Murphy, F., Ramakrishnan, V., Malkiewicz, A. et al. The role of modifications in codon discrimination by tRNALysUUU. Nat Struct Mol Biol 11, 1186–1191 (2004). https://doi.org/10.1038/nsmb861
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb861
This article is cited by
-
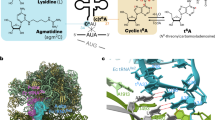
Structural insights into the decoding capability of isoleucine tRNAs with lysidine and agmatidine
Nature Structural & Molecular Biology (2024)
-
Threonine fuels glioblastoma through YRDC-mediated codon-biased translational reprogramming
Nature Cancer (2024)
-
A paralog of Pcc1 is the fifth core subunit of the KEOPS tRNA-modifying complex in Archaea
Nature Communications (2023)
-
Geometric alignment of aminoacyl-tRNA relative to catalytic centers of the ribosome underpins accurate mRNA decoding
Nature Communications (2023)
-
A prebiotically plausible scenario of an RNA–peptide world
Nature (2022)