Abstract
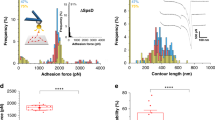
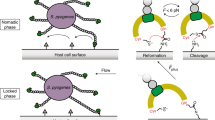
Although much progress has been made in the identification and characterization of adhesins borne by pathogenic bacteria, the molecular details underlying their interaction with host receptors remain largely unknown owing to the lack of appropriate probing techniques. Here we report a method, based on atomic force microscopy (AFM) with tips bearing biologically active molecules, for measuring the specific binding forces of individual adhesins and for mapping their distribution on the surface of living bacteria. First, we determined the adhesion forces between the heparin-binding haemagglutinin adhesin (HBHA) produced by Mycobacterium tuberculosis and heparin, used as a model sulphated glycoconjugate receptor. Both the adhesion frequency and adhesion force increased with contact time, indicating that the HBHA-heparin complex is formed via multiple intermolecular bridges. We then mapped the distribution of single HBHA molecules on the surface of living mycobacteria and found that the adhesin is not randomly distributed over the mycobacterial surface, but concentrated into nanodomains.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
22 June 2005
The website listed in the article has been modified
Notes
NOTE:The website listed in the article has been modified. The corrected citation should read: (F.D.M. and M.C. Vidal-Pessolani, unpublished data; http://genolist.pasteur.fr/Leproma/) The error has been corrected in the HTML version of the article, and this correction has been appended to the PDF version of the article.
References
Menozzi, F.D. et al. Identification of a heparin-binding hemagglutinin present in mycobacteria. J. Exp. Med. 184, 993–1001 (1996).
Pethe, K. et al. Characterization of the heparin-binding site of the mycobacterial heparin-binding hemagglutinin adhesin. J. Biol. Chem. 275, 14273–14280 (2000).
Reddy, V.M. & Kumar, B. Interaction of Mycobacterium avium complex with human respiratory epithelial cells. J. Infect. Dis. 181, 1189–1193 (2000).
Menozzi, F.D., Bischoff, R., Fort, E., Brennan, M.J. & Locht, C. Molecular characterization of the mycobacterial heparin-binding hemagglutinin, a mycobacterial adhesin. Proc. Natl. Acad. Sci. USA 95, 12625–12630 (1998).
Pethe, K. et al. The heparin-binding haemagglutinin of M. tuberculosis is required for extrapulmonary dissemination. Nature 412, 190–194 (2001).
Mueller-Ortiz, S.L. et al. Decreased infectivity despite unaltered C3 binding by a ΔhbhA mutant of Mycobacterium tuberculosis. Infect. Immun. 70, 6751–6760 (2002).
Delogu, G. & Brennan, M.J. Functional domains in the mycobacterial hemagglutinin, HBHA. J. Bacteriol. 181, 7464–7469 (1999).
Binnig, G., Quate, C.F. & Gerber, C. Atomic force microscope. Phys. Rev. Lett. 56, 930–933 (1986).
Engel, A. & Müller, D.J. Observing single biomolecules at work with the atomic force microscope. Nat. Struct. Biol. 7, 715–718 (2000).
Hörber, J.K. & Miles, M.J. Scanning probe evolution in biology. Science 302, 1002–1005 (2003).
Dufrêne, Y.F. Using nanotechniques to explore microbial surfaces. Nat. Rev. Microbiol. 2, 451–460 (2004).
Lee, G.U., Chrisey, L.A. & Colton, R.J. Direct measurement of the forces between complementary strands of DNA. Science 266, 771–773 (1994).
Hinterdorfer, P., Baumgartner, W., Gruber, H.J., Schilcher, K. & Schindler, H. Detection and localization of individual antibody-antigen recognition events by atomic force microscopy. Proc. Natl. Acad. Sci. USA 93, 3477–3481 (1996).
Lower, S.K., Hochella, M.F. & Beveridge, T.J. Bacterial recognition of mineral surfaces: nanoscale interactions between Shewanella and α-FeOOH. Science 292, 1360–1363 (2001).
Abu-Lail, N.I. & Camesano, T.A. Elasticity of Pseudomonas putida KT2442 surface polymers probed with single-molecule force microscopy. Langmuir 18, 4071–4081 (2002).
Ludwig, M., Dettmann, W. & Gaub, H.E. Atomic force microscope imaging contrast based on molecular recognition. Biophys. J. 72, 445–448 (1997).
Heinz, W.F. & Hoh, J.H. Spatially resolved force spectroscopy of biological surfaces using the atomic force microscope. Trends Biotechnol. 17, 143–150 (1999).
Grandbois, M., Dettmann, W., Benoit, M. & Gaub, H.E. Affinity imaging of red blood cells using an atomic force microscope. J. Histochem. Cytochem. 48, 719–724 (2000).
Lehenkari, P.P., Charras, G.T., Nykänen, A. & Horton, M.A. Adapting atomic force microscopy for cell biology. Ultramicroscopy 82, 289–295 (2000).
Almqvist, N. et al. Elasticity and adhesion force mapping reveals real-time clustering of growth factor receptors and associated changes in local cellular rheological properties. Biophys. J. 86, 1753–1762 (2004).
Luk, Y-Y. et al. Using liquid crystals to amplify protein-receptor interactions: design of surfaces with nanometer-scale topography that present histidine-tagged protein receptors. Langmuir 19, 1671–1680 (2003).
Rief, M., Oesterhelt, F., Heymann, B. & Gaub, H.E. Single molecule force spectroscopy on polysaccharides by atomic force microscopy. Science 275, 1295–1297 (1997).
Merkel, R., Nassoy, P., Leung, A., Ritchie, K. & Evans, E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature 397, 50–53 (1999).
Baumgartner, W. et al. Cadherin interaction probed by atomic force microscopy. Proc. Natl. Acad. Sci. USA 97, 4005–4010 (2000).
Auletta, T. β-Cyclodextrin host-guest complexes probed under thermodynamic equilibrium: thermodynamics and AFM force spectroscopy. J. Am. Chem. Soc. 126, 1577–1584 (2004).
Devadoss, P., Klegerman, M.E. & Groves, M.J. Surface morphology of Mycobacterium bovis BCG: relation to mechanisms of cellular aggregation. Microbios. 65, 111–125 (1991).
Florin, E.L., Moy, V.T. & Gaub, H.E. Adhesion forces between individual ligand-receptor pairs. Science 264, 415–417 (1994).
Margalit, H., Fischer, N. & Ben-Sasson, S.A. Comparative analysis of structurally defined heparin binding sequences reveals a distinct spatial distribution of basic residues. J. Biol. Chem. 268, 19228–19231 (1993).
Bernfield, M. et al. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 68, 729–777 (1999).
Tkachenko, E. & Simons, M. Clustering induces redistribution of syndecan-4 core protein into raft membrane domains. J. Biol. Chem. 277, 19946–19951 (2002).
Ofek, I., Hasty, D.L. & Sharon, N. Anti-adhesion therapy of bacterial diseases: prospects and problems. FEMS Immunol. Med. Microbiol. 38, 181–191 (2003).
Acknowledgements
This work was supported by the National Foundation for Scientific Research (FNRS), the Université Catholique de Louvain (Fonds Spéciaux de Recherche), the Federal Office for Scientific, Technical and Cultural Affairs (Interuniversity Poles of Attraction Programme), the Research Department of the Communauté Française de Belgique (Concerted Research Action), the INSERM, the Institut Pasteur de Lille and the Région Nord-Pas de Calais. N.L.A. and B.H.C. acknowledge support from the US National Science Foundation through the MRSEC program. We thank G. Delogù for the gift of E. coli BL21(pGD51), E. Pradel and M. Simonet for critical reading of the manuscript, L. Piraux for the use of the thermal evaporator, E. Ferain for the use of the scanning electron microscope and P. Hinterdorfer for stimulating discussions. Y.F.D. is a Research Associate of the FNRS.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Dupres, V., Menozzi, F., Locht, C. et al. Nanoscale mapping and functional analysis of individual adhesins on living bacteria. Nat Methods 2, 515–520 (2005). https://doi.org/10.1038/nmeth769
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth769
This article is cited by
-
Force spectroscopy of single cells using atomic force microscopy
Nature Reviews Methods Primers (2021)
-
Dual-pulse photoactivated atomic force microscopy
Scientific Reports (2021)
-
Use the force
Nature Physics (2020)
-
Development of a novel multiphysical approach for the characterization of mechanical properties of musculotendinous tissues
Scientific Reports (2019)
-
Glycan-mediated enhancement of reovirus receptor binding
Nature Communications (2019)