Abstract
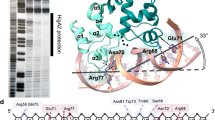
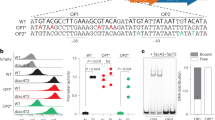
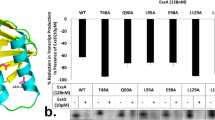
Conditional cooperativity is a common mechanism involved in transcriptional regulation of prokaryotic type II toxin–antitoxin operons and is intricately related to bacterial persistence. It allows the toxin component of a toxin–antitoxin module to act as a co-repressor at low doses of toxin as compared to antitoxin. When toxin level exceeds a certain threshold, however, the toxin becomes a de-repressor. Most antitoxins contain an intrinsically disordered region (IDR) that typically is involved in toxin neutralization and repressor complex formation. To address how the antitoxin IDR is involved in transcription regulation, we studied the phd–doc operon from bacteriophage P1. We provide evidence that the IDR of Phd provides an entropic barrier precluding full operon repression in the absence of Doc. Binding of Doc results in a cooperativity switch and consequent strong operon repression, enabling context-specific modulation of the regulatory process. Variations of this theme are likely to be a common mechanism in the autoregulation of bacterial operons that involve intrinsically disordered regions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Gerdes, K., Christensen, S.K. & Løbner-Olesen, A. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3, 371–382 (2005).
Helaine, S. et al. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343, 204–208 (2014).
Maisonneuve, E., Castro-Camargo, M. & Gerdes, K. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154, 1140–1150 (2013).
Garcia-Pino, A. et al. Allostery and intrinsic disorder mediate transcription regulation by conditional cooperativity. Cell 142, 101–111 (2010).
Loris, R. & Garcia-Pino, A. Disorder- and dynamics-based regulatory mechanisms in toxin-antitoxin modules. Chem. Rev. 114, 6933–6947 (2014).
Overgaard, M., Borch, J., Jørgensen, M.G. & Gerdes, K. Messenger RNA interferase RelE controls relBE transcription by conditional cooperativity. Mol. Microbiol. 69, 841–857 (2008).
Buts, L., Lah, J., Dao-Thi, M.H., Wyns, L. & Loris, R. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem. Sci. 30, 672–679 (2005).
De Gieter, S. et al. The intrinsically disordered domain of the antitoxin Phd chaperones the toxin Doc against irreversible inactivation and misfolding. J. Biol. Chem. 289, 34013–34023 (2014).
Lehnherr, H., Maguin, E., Jafri, S. & Yarmolinsky, M.B. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J. Mol. Biol. 233, 414–428 (1993).
McKinley, J.E. & Magnuson, R.D. Characterization of the Phd repressor-antitoxin boundary. J. Bacteriol. 187, 765–770 (2005).
Smith, J.A. & Magnuson, R.D. Modular organization of the Phd repressor/antitoxin protein. J. Bacteriol. 186, 2692–2698 (2004).
Kamada, K. & Hanaoka, F. Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol. Cell 19, 497–509 (2005).
Garcia-Pino, A., Zenkin, N. & Loris, R. The many faces of Fic: structural and functional aspects of Fic enzymes. Trends Biochem. Sci. 39, 121–129 (2014).
Garcia-Pino, A. et al. Doc of prophage P1 is inhibited by its antitoxin partner Phd through fold complementation. J. Biol. Chem. 283, 30821–30827 (2008).
Castro-Roa, D. et al. The Fic protein Doc uses an inverted substrate to phosphorylate and inactivate EF-Tu. Nat. Chem. Biol. 9, 811–817 (2013).
Anantharaman, V. & Aravind, L. New connections in the prokaryotic toxin-antitoxin network: relationship with the eukaryotic nonsense-mediated RNA decay system. Genome Biol. 4, R81 (2003).
Zhao, X. & Magnuson, R.D. Percolation of the phd repressor-operator interface. J. Bacteriol. 187, 1901–1912 (2005).
Magnuson, R., Lehnherr, H., Mukhopadhyay, G. & Yarmolinsky, M.B. Autoregulation of the plasmid addiction operon of bacteriophage P1. J. Biol. Chem. 271, 18705–18710 (1996).
Chan, W.T., Yeo, C.C., Sadowy, E. & Espinosa, M. Functional validation of putative toxin-antitoxin genes from the Gram-positive pathogen Streptococcus pneumoniae: phd-doc is the fourth bona-fide operon. Front. Microbiol. 5, 677 (2014).
Gazit, E. & Sauer, R.T. Stability and DNA binding of the phd protein of the phage P1 plasmid addiction system. J. Biol. Chem. 274, 2652–2657 (1999).
Bernadó, P., Mylonas, E., Petoukhov, M.V., Blackledge, M. & Svergun, D.I. Structural characterization of flexible proteins using small-angle X-ray scattering. J. Am. Chem. Soc. 129, 5656–5664 (2007).
Pelikan, M., Hura, G.L. & Hammel, M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen. Physiol. Biophys. 28, 174–189 (2009).
Magnuson, R. & Yarmolinsky, M.B. Corepression of the P1 addiction operon by Phd and Doc. J. Bacteriol. 180, 6342–6351 (1998).
Afif, H., Allali, N., Couturier, M. & Van Melderen, L. The ratio between CcdA and CcdB modulates the transcriptional repression of the ccd poison-antidote system. Mol. Microbiol. 41, 73–82 (2001).
De Jonge, N. et al. Rejuvenation of CcdB-poisoned gyrase by an intrinsically disordered protein domain. Mol. Cell 35, 154–163 (2009).
Monti, M.C. et al. Interactions of Kid-Kis toxin-antitoxin complexes with the parD operator-promoter region of plasmid R1 are piloted by the Kis antitoxin and tuned by the stoichiometry of Kid-Kis oligomers. Nucleic Acids Res. 35, 1737–1749 (2007).
Winther, K.S. & Gerdes, K. Regulation of enteric vapBC transcription: induction by VapC toxin dimer-breaking. Nucleic Acids Res. 40, 4347–4357 (2012).
Brown, B.L., Lord, D.M., Grigoriu, S., Peti, W. & Page, R. The Escherichia coli toxin MqsR destabilizes the transcriptional repression complex formed between the antitoxin MqsA and the mqsRA operon promoter. J. Biol. Chem. 288, 1286–1294 (2013).
Schumacher, M.A. et al. HipBA-promoter structures reveal the basis of heritable multidrug tolerance. Nature 524, 59–64 (2015).
Motlagh, H.N., Wrabl, J.O., Li, J. & Hilser, V.J. The ensemble nature of allostery. Nature 508, 331–339 (2014).
Hilser, V.J. & Thompson, E.B. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc. Natl. Acad. Sci. USA 104, 8311–8315 (2007).
Tzeng, S.R. & Kalodimos, C.G. Dynamic activation of an allosteric regulatory protein. Nature 462, 368–372 (2009).
Fuxreiter, M. et al. Disordered proteinaceous machines. Chem. Rev. 114, 6806–6843 (2014).
Ferreon, A.C., Ferreon, J.C., Wright, P.E. & Deniz, A.A. Modulation of allostery by protein intrinsic disorder. Nature 498, 390–394 (2013).
De la Cruz, M.A. et al. A toxin-antitoxin module of Salmonella promotes virulence in mice. PLoS Pathog. 9, e1003827 (2013).
Simanshu, D.K., Yamaguchi, Y., Park, J.H., Inouye, M. & Patel, D.J. Structural basis of mRNA recognition and cleavage by toxin MazF and its regulation by antitoxin MazE in Bacillus subtilis. Mol. Cell 52, 447–458 (2013).
Sterckx, Y.G. et al. An efficient method for the purification of proteins from four distinct toxin-antitoxin modules. Protein Expr. Purif. 108, 30–40 (2015).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
McCoy, A.J., Grosse-Kunstleve, R.W., Storoni, L.C. & Read, R.J. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 (2005).
Macke, T.J. & Case, D.A. Modeling unusual nucleic acid structures. in Molecular Modeling of Nucleic Acids 379–393 (American Chemical Society, Washington, DC, USA, 1998).
Bricogne, G. et al. BUSTER version 2.10.0 (Global Phasing Ltd, Cambridge, UK, 2011).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Davis, I.W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
Theobald, D.L. & Wuttke, D.S. THESEUS: maximum likelihood superpositioning and analysis of macromolecular structures. Bioinformatics 22, 2171–2172 (2006).
Konarev, P.V., Petoukhov, M.V., Volkov, V.V. & Svergun, D.I. ATSAS 2.1, a program package for small-angle scattering data analysis. J. Appl. Crystallogr. 39, 277–286 (2006).
Šali, A. & Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993).
Rambo, R.P. & Tainer, J.A. Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496, 477–481 (2013).
Houtman, J.C. et al. Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: application to adaptor protein complexes in cell signaling. Protein Sci. 16, 30–42 (2007).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Hohn, M. et al. SPARX, a new environment for Cryo-EM image processing. J. Struct. Biol. 157, 47–55 (2007).
Elmlund, H., Elmlund, D. & Bengio, S. PRIME: probabilistic initial 3D model generation for single-particle cryo-electron microscopy. Structure 21, 1299–1306 (2013).
Pettersen, E.F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Phillips, J.C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Acknowledgements
The research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7/2007-2013) under BioStruct-X (grant agreement no. 283570) and from the VIB, FWO-Vlaanderen, the Hercules Foundation, the Fonds National de Recherche Scientifique (FNRS) and the Fonds d'Encouragement à la Recherche ULB (FER-ULB). S.D.G. and A.G.-P. acknowledge the receipt of individual predoctoral and postdoctoral fellowships, respectively, from FWO-Vlaanderen. We also acknowledge the use of beamtime at the synchrotron beamlines BM29 (ESRF Grenoble, France) and SWING and PROXIMA1 (Soleil Gif-sur-Yvette, France) and thank the beamline staff for their support.
Author information
Authors and Affiliations
Contributions
A.G.-P. performed the calorimetry experiments; collected and analyzed crystallographic, SAXS and EM data; and wrote the paper and supervised the project. S.D.G. purified Doc and Phd, prepared the Phd–Oln1 crystals and collected SAXS data. A.T. ran and analyzed the MD simulations. H.D.G. prepared the Phd mutants. R.G.E. collected and analyzed EM data and calculated EM models. R.L. wrote the paper and supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–8 and Supplementary Tables 1–5. (PDF 34588 kb)
Rights and permissions
About this article
Cite this article
Garcia-Pino, A., De Gieter, S., Talavera, A. et al. An intrinsically disordered entropic switch determines allostery in Phd–Doc regulation. Nat Chem Biol 12, 490–496 (2016). https://doi.org/10.1038/nchembio.2078
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2078
This article is cited by
-
Programming conformational cooperativity to regulate allosteric protein-oligonucleotide signal transduction
Nature Communications (2023)
-
Type II toxin–antitoxin system in bacteria: activation, function, and mode of action
Biophysics Reports (2020)
-
Mechanism of regulation and neutralization of the AtaR–AtaT toxin–antitoxin system
Nature Chemical Biology (2019)
-
A dual role in regulation and toxicity for the disordered N-terminus of the toxin GraT
Nature Communications (2019)
-
A hidden competitive advantage of disorder
Nature (2017)