Abstract
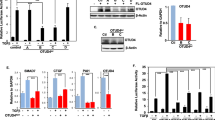
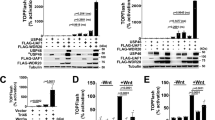
The TGFβ pathway is critical for embryonic development and adult tissue homeostasis. On ligand stimulation, TGFβ and BMP receptors phosphorylate receptor-activated SMADs (R-SMADs), which then associate with SMAD4 to form a transcriptional complex that regulates gene expression through specific DNA recognition1,2. Several ubiquitin ligases serve as inhibitors of R-SMADs3,4, yet no deubiquitylating enzyme (DUB) for these molecules has so far been identified. This has left unexplored the possibility that ubiquitylation of R-SMADs is reversible and engaged in regulating SMAD function, in addition to degradation5. Here we identify USP15 as a DUB for R-SMADs. USP15 is required for TGFβ and BMP responses in mammalian cells and Xenopus embryos. At the biochemical level, USP15 primarily opposes R-SMAD monoubiquitylation, which targets the DNA-binding domains of R-SMADs and prevents promoter recognition. As such, USP15 is critical for the occupancy of endogenous target promoters by the SMAD complex. These data identify an additional layer of control by which the ubiquitin system regulates TGFβ biology.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Wu, M. Y. & Hill, C. S. Tgf-β superfamily signaling in embryonic development and homeostasis. Dev. Cell 16, 329–343 (2009).
Wrana, J. L., Ozdamar, B., Le Roy, C. & Benchabane, H. Signaling Receptors of the TGF β family (Cold Spring Harbor Laboratory Press, 2008).
Itoh, S. & ten Dijke, P. Negative regulation of TGF-β receptor/Smad signal transduction. Curr. Opin. Cell Biol. 19, 176–184 (2007).
Lonn, P., Moren, A., Raja, E., Dahl, M. & Moustakas, A. Regulating the stability of TGFβ receptors and Smads. Cell Res. 19, 21–35 (2009).
Salmena, L. & Pandolfi, P. P. Changing venues for tumour suppression: balancing destruction and localization by monoubiquitylation. Nat. Rev. Cancer 7, 409–413 (2007).
Nijman, S. M. et al. A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773–786 (2005).
Dupont, S. et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFβ signaling, controls Smad4 monoubiquitination. Cell 136, 123–135 (2009).
Baker, R. T., Wang, X. W., Woollatt, E., White, J. A. & Sutherland, G. R. Identification, functional characterization, and chromosomal localization of USP15, a novel human ubiquitin-specific protease related to the UNP oncoprotein, and a systematic nomenclature for human ubiquitin-specific proteases. Genomics 59, 264–274 (1999).
Miyazono, K., Maeda, S. & Imamura, T. Coordinate regulation of cell growth and differentiation by TGF-β superfamily and Runx proteins. Oncogene 23, 4232–4237 (2004).
Akhurst, R. J. & Derynck, R. TGF-β signaling in cancer—a double-edged sword. Trends Cell Biol. 11, S44–S51 (2001).
Niehrs, C. Regionally specific induction by the Spemann–Mangold organizer. Nat. Rev. Genet. 5, 425–434 (2004).
De Robertis, E. M., Larrain, J., Oelgeschlager, M. & Wessely, O. The establishment of Spemann’s organizer and patterning of the vertebrate embryo. Nat. Rev. Genet. 1, 171–181 (2000).
Fuentealba, L. C. et al. Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell 131, 980–993 (2007).
Zhu, H., Kavsak, P., Abdollah, S., Wrana, J. L. & Thomsen, G. H. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 400, 687–693 (1999).
Sapkota, G., Alarcon, C., Spagnoli, F. M., Brivanlou, A. H. & Massague, J. Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol. Cell 25, 441–454 (2007).
Hill, C. S. Nucleocytoplasmic shuttling of Smad proteins. Cell Res. 19, 36–46 (2009).
Stroschein, S. L., Wang, W., Zhou, S., Zhou, Q. & Luo, K. Negative feedback regulation of TGF-β signaling by the SnoN oncoprotein. Science 286, 771–774 (1999).
Shi, Y. Structural insights on Smad function in TGFβ signaling. Bioessays 23, 223–232 (2001).
Chai, J. et al. Features of a Smad3 MH1–DNA complex. Roles of water and zinc in DNA binding. J. Biol. Chem. 278, 20327–20331 (2003).
Maspero, E. et al. Structure of the HECT:ubiquitin complex and its role in ubiquitin chain elongation. EMBO Rep. 12, 342–349 (2011).
Chen, X., Rubock, M. J. & Whitman, M. A transcriptional partner for MAD proteins in TGF-β signalling. Nature 383, 691–696 (1996).
Inman, G. J., Nicolas, F. J. & Hill, C. S. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-β receptor activity. Mol. Cell 10, 283–294 (2002).
Dai, F., Lin, X., Chang, C. & Feng, X. H. Nuclear export of Smad2 and Smad3by RanBP3 facilitates termination of TGF-β signaling. Dev. Cell 16, 345–357 (2009).
Alarcon, C. et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-β pathways. Cell 139, 757–769 (2009).
Dupont, S. et al. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell 121, 87–99 (2005).
Morsut, L. et al. Negative control of Smad activity by ectodermin/Tif1γ patterns the mammalian embryo. Development 137, 2571–2578 (2010).
Fukuda, M., Gotoh, I., Adachi, M., Gotoh, Y. & Nishida, E. A novel regulatory mechanism in the mitogen-activated protein (MAP) kinase cascade. Role ofnuclear export signal of MAP kinase kinase. J. Biol. Chem. 272, 32642–32648 (1997).
Levy, L. et al. Arkadia activates Smad3/Smad4-dependent transcription by triggering signal-induced SnoN degradation. Mol. Cell Biol. 27, 6068–6083 (2007).
Cordenonsi, M. et al. Integration of TGF-β and Ras/MAPK signaling through p53 phosphorylation. Science 315, 840–843 (2007).
Martello, G. et al. MicroRNA control of Nodal signalling. Nature 449, 183–188 (2007).
Yu, P. B. et al. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat. Chem. Biol. 4, 33–41 (2008).
Adorno, M. et al. A Mutant-p53/Smad complex opposes p63 to empower TGFβ-induced metastasis. Cell 137, 87–98 (2009).
Gao, S. et al. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-β signaling. Mol. Cell 36, 457–468 (2009).
Cordenonsi, M et al. Links between tumor suppressors: p53 is required for TGF-β gene responses by cooperating with Smads. Cell 113, 301–314 (2003).
Fontemaggi, G. et al. The transcriptional repressor ZEB regulates p73 expression at the crossroad between proliferation and differentiation. Mol. Cell Biol. 21, 8461–8470 (2001).
Acknowledgements
We thank W. Dubiel, P. ten Dijke, C. Hill, A. Moustakas and E. De Robertis for gifts of reagents, and E. Maspero and G. Fontemaggi for help with enzyme purification and ChIP. We are grateful to M. Aragona, F. Zanconato and O. Wessely for comments on the manuscript. This work is supported by grants from the following agencies: AIRC (Italian Association for Cancer Research) PI and AIRC Special Program Molecular Clinical Oncology ‘5 per mille’, University of Padua strategic grant, excellence grant IIT and Comitato Promotore Telethon grant to S.P., and AIRC and MIUR grants to S.D. and M.C. M.I. was a recipient of a Toyobo and Uehara Memorial Foundations grant and is a Marie Curie FP-7 IIF fellow.
Author information
Authors and Affiliations
Contributions
M.I. and S.P. designed research; M.I. and A. Manfrin carried out all the biochemical and functional assays in cells and Xenopus embryos. A. Mamidi and S.D. purified SMADs and carried out in vitro binding assays; G.M., S.S. and M.C. helped with Xenopus assays; L.M. and E.E. helped with mesenchymal stem cells and immunofluorescence; S.M. carried out the modelling analysis; S.P. prepared recombinant E3 ligases; S.P. coordinated the work; M.I. and S.P. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1102 kb)
Rights and permissions
About this article
Cite this article
Inui, M., Manfrin, A., Mamidi, A. et al. USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nat Cell Biol 13, 1368–1375 (2011). https://doi.org/10.1038/ncb2346
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2346
This article is cited by
-
Targeting ubiquitin specific proteases (USPs) in cancer immunotherapy: from basic research to preclinical application
Journal of Experimental & Clinical Cancer Research (2023)
-
Downregulation of Ubiquitin-Specific Protease 15 (USP15) Does Not Provide Therapeutic Benefit in Experimental Mesial Temporal Lobe Epilepsy
Molecular Neurobiology (2023)
-
USP15 regulates p66Shc stability associated with Drp1 activation in liver ischemia/reperfusion
Cell Death & Disease (2022)
-
Positive feedback regulation between USP15 and ERK2 inhibits osteoarthritis progression through TGF-β/SMAD2 signaling
Arthritis Research & Therapy (2021)
-
Deubiquitylases in developmental ubiquitin signaling and congenital diseases
Cell Death & Differentiation (2021)