Abstract
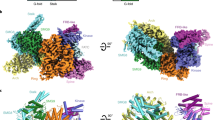
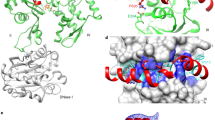
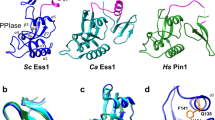
Symplekin (Pta1 in yeast) is a scaffold in the large protein complex that is required for 3′-end cleavage and polyadenylation of eukaryotic messenger RNA precursors (pre-mRNAs)1,2,3,4; it also participates in transcription initiation and termination by RNA polymerase II (Pol II)5,6. Symplekin mediates interactions between many different proteins in this machinery1,2,7,8,9, although the molecular basis for its function is not known. Here we report the crystal structure at 2.4 Å resolution of the amino-terminal domain (residues 30–340) of human symplekin in a ternary complex with the Pol II carboxy-terminal domain (CTD) Ser 5 phosphatase Ssu72 (refs 7, 10–17) and a CTD Ser 5 phosphopeptide. The N-terminal domain of symplekin has the ARM or HEAT fold, with seven pairs of antiparallel α-helices arranged in the shape of an arc. The structure of Ssu72 has some similarity to that of low-molecular-mass phosphotyrosine protein phosphatase18,19, although Ssu72 has a unique active-site landscape as well as extra structural features at the C terminus that are important for interaction with symplekin. Ssu72 is bound to the concave face of symplekin, and engineered mutations in this interface can abolish interactions between the two proteins. The CTD peptide is bound in the active site of Ssu72, with the pSer 5-Pro 6 peptide bond in the cis configuration, which contrasts with all other known CTD peptide conformations20,21. Although the active site of Ssu72 is about 25 Å from the interface with symplekin, we found that the symplekin N-terminal domain stimulates Ssu72 CTD phosphatase activity in vitro. Furthermore, the N-terminal domain of symplekin inhibits polyadenylation in vitro, but only when coupled to transcription. Because catalytically active Ssu72 overcomes this inhibition, our results show a role for mammalian Ssu72 in transcription-coupled pre-mRNA 3′-end processing.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Takagaki, Y. & Manley, J. L. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol. Cell. Biol. 20, 1515–1525 (2000)
Zhao, J., Kessler, M. M., Helmling, S., O’Connor, J. P. & Moore, C. L. Pta1, a component of yeast CFII, is required for both cleavage and poly(A) addition of mRNA precursor. Mol. Cell. Biol. 19, 7733–7740 (1999)
Zhao, J., Hyman, L. & Moore, C. L. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63, 405–445 (1999)
Mandel, C. R., Bai, Y. & Tong, L. Protein factors in pre-mRNA 3′-end processing. Cell. Mol. Life Sci. 65, 1099–1122 (2008)
Calvo, O. & Manley, J. L. Strange bedfellows: polyadenylation factors at the promoter. Genes Dev. 17, 1321–1327 (2003)
Moore, M. J. & Proudfoot, N. J. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136, 688–700 (2009)
He, X. et al. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes Dev. 17, 1030–1042 (2003)
Nedea, E. et al. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J. Biol. Chem. 278, 33000–33010 (2003)
Ghazy, M. A., He, X., Singh, B. N., Hampsey, M. & Moore, C. The essential N terminus of the Pta1 scaffold protein is required for snoRNA transcription termination and Ssu72 function but is dispensible for pre-mRNA 3′-end processing. Mol. Cell. Biol. 29, 2296–2307 (2009)
Dichtl, B. et al. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell 10, 1139–1150 (2002)
Meinhart, A., Silberzahn, T. & Cramer, P. The mRNA transcription/processing factor Ssu72 is a potential tyrosine phosphatase. J. Biol. Chem. 278, 15917–15921 (2003)
Ganem, C. et al. Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast. EMBO J. 22, 1588–1598 (2003)
Krishnamurthy, S., He, X., Reyes-Reyes, M., Moore, C. L. & Hampsey, M. Ssu72 is an RNA polymerase II CTD phosphatase. Mol. Cell 14, 387–394 (2004)
Hausmann, S., Koiwa, H., Krishnamurthy, S., Hampsey, M. & Shuman, S. Different strategies for carboxyl-terminal domain (CTD) recognition by serine 5-specific CTD phosphatases. J. Biol. Chem. 280, 37681–37688 (2005)
St-Pierre, B. et al. Conserved and specific functions of mammalian ssu72. Nucleic Acids Res. 33, 464–477 (2005)
Reyes-Reyes, M. & Hampsey, M. Role for the Ssu72 C-terminal domain phosphatase in RNA polymerase II transcription elongation. Mol. Cell. Biol. 27, 926–936 (2007)
Ansari, A. & Hampsey, M. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 19, 2969–2978 (2005)
Su, X.-D., Taddei, N., Stefani, M., Ramponi, G. & Nordlund, P. The crystal structure of a low-molecular-weight phosphotyrosine protein phosphatase. Nature 370, 575–578 (1994)
Zhang, M., Van Etten, R. L. & Stauffacher, C. V. Crystal structure of bovine heart phosphotyrosyl phosphatase at 2.2-Å resolution. Biochemistry 33, 11097–11105 (1994)
Meinhart, A., Kamenski, T., Hoeppner, S., Baumli, S. & Cramer, P. A structural perspective of CTD function. Genes Dev. 19, 1401–1415 (2005)
Zhang, Y. et al. Determinants for dephosphorylation of the RNA polymerase II C-terminal domain by Scp1. Mol. Cell 24, 759–770 (2006)
Kennedy, S. A. et al. Crystal structure of the HEAT domain from the pre-mRNA processing factor symplekin. J. Mol. Biol. 392, 115–128 (2009)
Xu, Y.-X., Hirose, Y., Zhou, X. Z., Lu, K. P. & Manley, J. L. Pin1 modulates the structure and function of human RNA polymerase II. Genes Dev. 17, 2765–2776 (2003)
Krishnamurthy, S., Ghazy, M. A., Moore, C. & Hampsey, M. Functional interaction of the Ess1 prolyl isomerase with components of the RNA polymerase II initiation and termination machineries. Mol. Cell. Biol. 29, 2925–2934 (2009)
Singh, N. et al. The Ess1 prolyl isomerase is required for transcription termination of small noncoding RNAs via the Nrd1 pathway. Mol. Cell 36, 255–266 (2009)
Hirose, Y. & Manley, J. L. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395, 93–96 (1998)
Rozenblatt-Rosen, O. et al. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc. Natl Acad. Sci. USA 106, 755–760 (2009)
Shi, Y. et al. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol. Cell 33, 365–376 (2009)
Takagaki, Y., Ryner, L. C. & Manley, J. L. Separation and characterization of a poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA polyadenylation. Cell 52, 731–742 (1988)
McCracken, S. et al. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385, 357–361 (1997)
Hendrickson, W. A., Horton, J. R. & LeMaster, D. M. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 9, 1665–1672 (1990)
Hendrickson, W. A. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science 254, 51–58 (1991)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Weeks, C. M. & Miller, R. The design and implementation of SnB v2.0. J. Appl. Cryst. 32, 120–124 (1999)
Terwilliger, T. C. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 374, 22–37 (2003)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Emsley, P. & Cowtan, K. D. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004)
Brunger, A. T. et al. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997)
Jogl, G., Tao, X., Xu, Y. & Tong, L. COMO: A program for combined molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 57, 1127–1134 (2001)
Hirose, Y. & Manley, J. L. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395, 93–96 (1998)
Chapman, R. D. et al. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 318, 1780–1782 (2007)
Acknowledgements
We thank F. Forouhar and J. Seetharaman for help with data collection; T. Auperin for initial experiments with symplekin; R. Abramowitz and J. Schwanof for setting up the X4C beamline; S. Myers for setting up the X29A beamline at the National Synchrotron Light Source; M. Heidemann and D. Eick for providing the 3E10 antibody; N. Rao and C. Logan for HeLa nuclear extract and help with in vitro assays; and J. Decatur for characterizing the phosphopeptide by NMR. This research is supported in part by grants from the National Institutes of Health to L.T. (GM077175) and J.L.M. (GM028983).
Author information
Authors and Affiliations
Contributions
K.X., S.X., T.K. and M.M.B. performed protein expression, purification and crystallization experiments. K.X., S.X. and L.T. conducted crystallographic data collection, structure determination and refinement. T.N. and K.X. performed polyadenylation experiments. K.X. performed Ssu72 phosphatase assays. All authors commented on the manuscript. L.T. and J.L.M. designed the experiments, analysed the data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Text, Supplementary Table 1 and Supplementary Figures 1-11 with legends. (PDF 3949 kb)
Rights and permissions
About this article
Cite this article
Xiang, K., Nagaike, T., Xiang, S. et al. Crystal structure of the human symplekin–Ssu72–CTD phosphopeptide complex. Nature 467, 729–733 (2010). https://doi.org/10.1038/nature09391
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09391
This article is cited by
-
Driving forces behind phase separation of the carboxy-terminal domain of RNA polymerase II
Nature Communications (2023)
-
Context-specific regulation and function of mRNA alternative polyadenylation
Nature Reviews Molecular Cell Biology (2022)
-
Diverse and conserved roles of the protein Ssu72 in eukaryotes: from yeast to higher organisms
Current Genetics (2021)
-
Drosophila melanogaster retrotransposon and inverted repeat-derived endogenous siRNAs are differentially processed in distinct cellular locations
BMC Genomics (2017)
-
Structure of the cohesin loader Scc2
Nature Communications (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.