Abstract
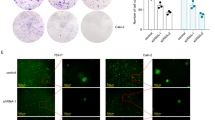
The aim of this study was to investigate the role and mechanism of miR-23a in the regulation of BCR/ABL and to provide a new prognostic biomarker for chronic myeloid leukemia (CML). The expression levels of miR-23a and BCR/ABL were assessed in 42 newly diagnosed CML patients, 37 CML patients in first complete remission and 25 healthy controls. Quantitative real-time PCR, western blot analysis and colony formation assay were used to evaluate changes induced by overexpression or inhibition of miR-23a or BCR/ABL. MiR-23a mimic or negative control mimic was transfected into a CML cell line (K562) and two lung cancer cell lines (H157 and SKMES1) using Lipofectamine 2000, and the cells were used for real-time reverse transcription-PCR (RT-PCR) and western blot analysis. We found that the downregulation of miR-23a expression was a frequent event in both leukemia cell lines and primary leukemic cells from patients with de novo CML. The microarray results showed that most of the CML patients expressed high levels of BCR/ABL and low levels of miR-23a. Real-time RT-PCR and western blot analysis showed that the BCR/ABL levels in miR-23a-transfected cells were lower than those in the control groups. Ectopic expression of miR-23a in K562 cells led to cellular senescence. Moreover, when K562 cells were treated with 5-aza-2′-deoxycytidine, a DNA methylation inhibitor, BCR/ABL expression was upregulated, which indicates epigenetic silencing of miR-23a in leukemic cells. BCR/ABL and miR-23a expressions were inversely related to CML, and BCR/ABL expression was regulated by miR-23a in leukemic cells. The epigenetic silencing of miR-23a led to derepression of BCR/ABL expression, and consequently contributes to CML development and progression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Teichler S, Illmer T, Roemhild J, Ovcharenko D, Stiewe T, Neubauer A. MicroRNA29a regulates the expression of the nuclear oncogene Ski. Blood 2011; 118: 1899–1902.
Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS . A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 2003; 9: 1274–1281.
Jørgensen HG, Copland M, Allan EK, Jiang X, Eaves A, Eaves C . Intermittent exposure of primitive quiescent chronic myeloid leukemia cells to granulocyte-colony stimulating factor in vitro promotes their elimination by imatinib mesylate. Clin Cancer Res 2006; 12: 626–633.
Ries C, Pitsch T, Mentele R, Zahler S, Egea V, Nagase H et al. Identification of a novel 82 kDa proMMP-9 species associated with the surface of leukaemic cells: (auto-)catalytic activation and resistance to inhibition by TIMP-1. Biochem J 2007; 405: 547–558.
Yu Q, Stamenkovic I . Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-β and promotes tumor invasion and angiogenesis. Genes Dev 2000; 14: 163–176.
Fridman R, Toth M, Chvyrkova I, Meroueh S, Mobashery S . Cell surface association of matrix metalloproteinase-9 (gelatinase B). Cancer Metast Rev 2003; 22: 153–166.
Stefanidakis M, Koivunen E . Cell-surface association between matrix metalloproteinases and integrins: role of the complexes in leukocyte migration and cancer progression. Blood 2006; 108: 1441–1450.
Xishan Z, Xinna Z, Baoxin H, Jun R . Impaired immunomodulatory function of chronic myeloid leukemia cancer stem cells and the possible mechanism involved in it. Cancer Immunol Immunother 2013; 62: 689–703.
Redondo-Muñoz J, Escobar-Díaz E, Samaniego R . MMP-9 in B-cell chronic lymphocytic leukemia is up-regulated by alpha4beta1 integrin or CXCR4 engagement via distinct signaling pathways, localizes to podosomes, and is involved in cell invasion and migration. Blood 2006; 108: 3143–3151.
Zhu X, Lin Z, Du J, Zhou X, Yang L, Liu G . Studies on microRNAs that are correlated with the cancer stem cells in chronic myeloid leukemia. Mol Cell Biochem 2014; 390: 75–84.
Janowska-Wieczorek A, Majka M, Marquez-Curtis L, Wertheim JA, Turner AR, Ratajczak MZ . Bcr-abl-positive cells secrete angiogenic factors including matrix metalloproteinases and stimulate angiogenesis in vivo in Matrigel implants. Leukemia 2002; 16: 1160–1166.
Chen Z, Chen L, Dai H, Wang P, Gao S, Wang K . MiR-301a promotes pancreatic cancer cell proliferation by directly inhibiting bim expression. J Cell Biochem 2012; 113: 3229–3235.
Lu Z, Li Y, Takwi A, Li B, Zhang J, Conklin DJ et al. miR-301a as an NF-kappaB activator in pancreatic cancer cells. EMBO J 2011; 30: 57–67.
Shishodia S, Sethi G, Konopleva M, Andreeff M, Aggarwal BB . A synthetic triterpenoid, CDDO-Me, inhibits IkappaBalpha kinase and enhances apoptosis induced by TNF and chemotherapeutic agents through down-regulation of expression of nuclear factor kappaB-regulated gene products in human leukemic cells. Clin Cancer Res 2006; 12: 1828–1838.
Kaneta Y, Kagami Y, Tsunoda T, Ohno R, Nakamura Y, Katagiri T . Genome-wide analysis of gene-expression profiles in chronic myeloid leukemia cells using a cDNA microarray. Int J Oncol 2003; 23: 681–691.
MacLellan SA, MacAulay C, Lam S, Garnis C . Pre-profiling factors influencing serum microRNA levels. BMC Clin Pathol 2014; 21: 14–27.
Wang X, Tian L, Wu H, Jiang X, Du L, Zhang H et al. Expression of miRNA-130a in nonsmall cell lung cancer. Am J Med Sci 2010; 340: 385–388.
Chen Y, Gorski DH . Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood 2008; 111: 1217–1226.
Suresh S, McCallum L, Lu W, Lazar N, Perbal B, Irvine AE . microRNAs 130a/b are regulated by BCR-ABL and downregulate expression of CCN3 in CML. J Cell Commun Signal 2011; 5: 183–191.
Zhou P, Jiang W, Wu L, Chang R, Wu K, Wang Z . miR-301a is a candidate oncogene that targets the homeobox gene Gax in human hepatocellular carcinoma. Dig Dis Sci 2012; 57: 1171–1180.
Shi W, Gerster K, Alajez NM, Tsang J, Waldron L, Pintilie M et al. MicroRNA-301 mediates proliferation and invasion in human breast cancer. Cancer Res 2011; 71: 2926–2937.
Kovaleva V, Mora R, Park YJ, Plass C, Chiramel AI, Bartenschlager R et al. MiRNA-130a targets ATG2B and DICER1 to inhibit autophagy and trigger killing of chronic lymphocytic leukemia cells. Cancer Res 2012; 72: 1763–1772.
Patel N, Tahara SM, Malik P, Kalra VK . Involvement of miR-30c and miR-301a in immediate induction of plasminogen activator inhibitor-1 by placental growth factor in human pulmonary endothelial cells. Biochem J 2011; 434: 473–482.
Baskerville S, Bartel DP . Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 2005; 11: 241–247.
Pouladi N, Kouhsari SM, Feizi MH, Gavgani RR, Azarfam P . Overlapping region of p53/Wrap53 transcripts: mutational analysis and sequence similarity with microRNA-4732-5p. Asian Pac J Cancer Prev 2013; 14: 3503–3507.
Wang L, Li B, Li L, Wang T . microRNA-497 suppresses proliferation and induces apoptosis in prostate cancer cells. Asian Pac J Cancer Prev 2013; 14: 3499–3502.
Xing HJ, Li YJ, Ma QM, Wang AM, Wang JL, Sun M et al. Identification of microRNAs present in congenital heart disease associated copy number variants. Eur Rev Med Pharmacol Sci 2013; 17 2114–2120.
Li X, Zhang X, Wang T, Sun C, Jin T, Yan H et al. Regulation by bisoprolol for cardiac microRNA expression in a rat volume-overload heart failure model. J Nanosci Nanotechnol 2013; 13: 5267–5275.
Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA 2005; 102: 13944–13949.
Cheng AM, Byrom MW, Shelton J, Ford LP . Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res 2005; 33: 1290–1297.
Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A et al. microRNA-responsive 'sensor' transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet 2004; 36: 1079–1083.
Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci USA 2005; 102: 18081–18086.
Karaayvaz M, Zhai H, Ju J . miR-129 promotes apoptosis and enhances chemosensitivity to 5-fluorouracil in colorectal cancer. Cell Death Dis 2013; 4: e659.
Zhang J, Zhang D, Wu GQ, Feng ZY, Zhu SM . Propofol inhibits the adhesion of hepatocellular carcinoma cells by upregulating microRNA-199a and downregulating MMP-9 expression. Hepatobiliary Pancreat Dis Int 2013; 12: 305–309.
Ma D, Tao X, Gao F, Fan C, Wu D . MiR-224 functions as an onco-miRNA in hepatocellular carcinoma cells by activating AKT signaling. Oncol Lett 2012; 4: 483–488.
Goedeke L, Vales-Lara FM, Fenstermaker M, Cirera-Salinas D, Chamorro-Jorganes A, Ramírez CM et al. A regulatory role for microRNA 33* in controlling lipid metabolism gene expression. Mol Cell Biol 2013; 33 2339–2352.
Cheng C, Li W, Zhang Z, Yoshimura S, Hao Q, Zhang C et al. MicroRNA-144 is regulated by activator protein-1 (AP-1) and decreases expression of Alzheimer disease-related a disintegrin and metalloprotease 10 (ADAM10). J Biol Chem 2013; 288: 13748–13761.
Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z et al. A combined computational experimental approach predicts human microRNA targets. Genes Dev 2004; 18: 1165–1178.
Krek A . Combinatorial microRNA target predictions. Nature Genet 2005; 37: 495–500.
Grun D, Wang Y, Langenberger D, Gunsalus KC, Rajewsky N . MicroRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput Biol 2005; 1: e13.
Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P et al. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol 2004; 5: R68.
Acknowledgements
The research was funded by the National Science Foundation (81100366) and Beijing Nova Programme (2013041).
Author Contributions
ZX devised the experiments and performed biologic assays. LZ performed the molecular assays. Li Xianjun and Cao Guangxin helped to revise the manuscript. Li Xianjun performed statistical analysis and Liu Gang reviewed all experimental data for the entire study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Xishan, Z., Xianjun, L., Ziying, L. et al. The malignancy suppression role of miR-23a by targeting the BCR/ABL oncogene in chromic myeloid leukemia. Cancer Gene Ther 21, 397–404 (2014). https://doi.org/10.1038/cgt.2014.44
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cgt.2014.44
This article is cited by
-
Role of miRNAs to control the progression of Chronic Myeloid Leukemia by their expression levels
Medical Oncology (2024)
-
MicroRNAs as prognostic biomarker and relapse indicator in leukemia
Clinical and Translational Oncology (2017)
-
Clinical significance of microRNAs in chronic and acute human leukemia
Molecular Cancer (2016)
-
High expression of MAP7 predicts adverse prognosis in young patients with cytogenetically normal acute myeloid leukemia
Scientific Reports (2016)
-
miR clusters target cellular functional complexes by defining their degree of regulatory freedom
Cancer and Metastasis Reviews (2016)