Abstract
To analyse the phenotype of breast tumours that express oestrogen receptor-β (ERβ) alone tissue microarrays were used to investigate if ERβ isoforms are associated with specific prognostic markers and gene expression phenotypes in ERα-negative tumours. ERα-negative tumours were positive for ERβ1 in 58% of cases (n=122/210), total ERβ in 60% (n=115/192) and ERβ2/cx in 57% of cases (n=114/199). Oestrogen receptor-β1 and total ERβ were significantly correlated with Ki67 (r=0.28, P<0.0001, n=209; r=0.29, P<0.0001, n=191) and with CK5/6, a marker of the basal phenotype (r=0.20, P=0.0106, n=170; r=0.18, P=0.0223, n=158). ERβ2/cx was strongly associated with p-c-Jun and NF-κBp65 (r=0.53, P<0.0001, n=93; r=0.35, P<0.0001, n=176). This study shows that a range of ERβ isoform expression occurs in ERα-negative breast tumours. While expression of ERβ1, total and ERβ2/cx are correlated, individual forms show associations with certain phenotypes that suggest different roles in subsets of ERα-negative cancers. Based on our in vivo observations, ERβ may have the potential to become a therapeutic target in the specific subcohort of ERα-negative breast cancers.
Similar content being viewed by others
Main
Oestrogen receptor-α (ERα) is an important biomarker of response to endocrine therapy in breast cancer (Osborne, 1998). However, the definition of ER status in breast cancer is potentially more complex, since there are now two known ERs, ERα and ERβ. Oestrogen receptor-β is expressed in both normal and neoplastic human breast tissue (Leygue et al, 1998; Mann et al, 2001; Murphy et al, 2002; Fuqua et al, 2003; Skliris et al, 2003) but its role in either tissue remains unknown. Several isoforms of ERβ have been identified, which are either exon deletions or products of alternative splicing which result in proteins that are truncated at the C-terminus and do not bind ligand (Lu et al, 1998; Ogawa et al, 1998; Fuqua et al, 1999; Leygue et al, 1999; Saunders et al, 2002). Thirty percent of breast tumours are classified as ER negative at the time of diagnosis and will be mostly resistant to endocrine therapy (Lapidus et al, 1998; Osborne, 1998). However, the previous assays used for ER measurement favoured the detection of ERα (Harvey et al, 1999; Brouillet et al, 2001) and we now know that some of these tumours express ERβ (Murphy et al, 2003). Considering studies where ERβ protein expression was determined, the pooled data sets were used to estimate the frequency of ERβ and ERα status in breast cancers (Murphy et al, 2003). The most frequently occurring tumour type is ERα+/ERβ+ (∼60%) with similar frequencies of the other three ER phenotypes (ERα+/ERβ−; ERα−/ERβ+; ERα−/ERβ−) at 10–20% (Murphy et al, 2003). It is important to note that there are two groups of ERβ-positive breast tumours, those with coexpression of ERα and those expressing ERβ alone. The former is the most frequent and probably dominates the analysis of most previously reported correlative studies, and hence the positive association of ERβ expression generally with good prognosis and good clinical outcome with respect to tamoxifen treatment (Mann et al, 2001; Omoto et al, 2001; Murphy et al, 2002; Iwase et al, 2003; Esslimani-Sahla et al, 2004; Fleming et al, 2004; Hopp et al, 2004; Myers et al, 2004; Nakopoulou et al, 2004). There is little data exploring tumours that express ERβ alone. Under the current system of determining ER status, these are classified clinically as ER negative, and currently there are few markers for further subclassifying these ERα-negative cancers. Nevertheless recent data show that some invasive breast cancers expressing the basal cytokeratin CK5/6, may represent one ERα-negative subset, known as the basal epithelial phenotype and show a relatively poor prognosis (Perou et al, 2000; Sorlie et al, 2001, 2003; Nielsen et al, 2004). In the present study, we have investigated the level and frequency of expression of ERβ in ER-negative tumours and its association with the basal phenotype and other established markers of prognosis, such as indicators of signal transduction pathways, proliferative and apoptotic markers.
Materials and methods
Tissues
All invasive breast cancers used in the current study were obtained from the Manitoba Breast Tumour Bank (MBTB, Department of Pathology, University of Manitoba) (Watson et al, 1996), which operates with approval from the Faculty of Medicine, University of Manitoba, Research Ethics Board. All samples included in the MBTB are rapidly frozen at −70°C immediately after surgical removal. A portion of the frozen tissue from each case is then processed to create matched formalin-fixed paraffin-embedded and frozen tissue blocks.
Clinical–pathological characteristics of the patient cohort
Cases selected for this study were on the basis of (a) minimum patient follow-up of 36 months, (b) invasive components occupying more than 20% of the tumour section, while normal epithelial areas comprised no more than 10% of the epithelial content and (c) ER-negative status as defined by ligand binding analysis (LBA) of ⩽3 fmol mg−1 protein. The criteria for interpretation of the variables were as follows: (a) PR-positive status was defined as >15 fmol mg−1 protein by LBA; (b) grade, (Nottingham system), was assigned to low (scores 3–5), moderate (scores 6 and 7), or high (scores 8 and 9) categories; (c) tumour size, was assigned either small (⩽2 cm) or large (>2 cm) categories; (d) tumour inflammation was assessed by a scale from 1 to 5 and then assigned to low (scores 1–3) or high (scores 4 and 5) categories. All patients were treated with surgery and for 29 patients this was the only treatment regimen. The remaining patients received a variety of additional treatments, hormonal therapy (28), chemotherapy (49) or radiotherapy (9) alone, or combination of radiation followed by hormonal therapy (8), hormonal and chemotherapy (16), hormonal and chemotherapy (19) or chemotherapy (46), and for 6 patients the treatment regime was unknown.
Tissue microarrays
The histopathology of all MBTB cases has been assessed and entered into a computerised database to enable selection based on composition of the tissue as well as clinical–pathological parameters. After selection, cases were rereviewed on H&E sections by a breast histopathologist (PHW). Tissue microarrays (TMAs) from a total cohort of 255 ERα negative (ERα–255TMA), primary invasive ductal breast carcinomas were constructed. Briefly, duplicate core tissue samples (0.6 mm diameter), were taken from selected areas of maximum cellularity for each tumour with a tissue arrayer instrument (Beecher Instruments, Silver Spring, MD, USA). Although the TMA consisted of 255 cases of ER-negative tumours as determined by LBA (ER+ >3 fmol mg−1 protein), 39 of these were subsequently found to be ERα+ by immunohistochemistry (IHC) and were excluded from the later analysis.
Immunohistochemical assay
Serial sections (5 μm) of the ERα–255TMA were cut, mounted on Fisherbrand Superfrost/plus slides (Fisher Scientific, USA) and stained using IHC with commercially available specific antibodies (Table 1). Further details of the three specific ERβ antibodies are as follows: ERβ1 (polyclonal, GC17/385P, Biogenex, CA, USA, raised to peptide containing amino acids 449–465) at 1 : 100 dilution; total ERβ (monoclonal, 14C8, Genetex, TX, USA, raised to peptide containing amino acids 1–153) at 1 : 100; ERβ2/cx (mouse monoclonal, clone 57/3, raised to synthetic peptide derived from the specific C-terminus of hERβ2/cx isoform; Serotec, UK) used at 1 : 20. Briefly, sections were dewaxed in two xylene baths (5 min each), taken through a series of alcohols (100, 95, 70%), rehydrated in distilled water and then submitted to heat-induced antigen retrieval for 8 min in the presence of a citrate buffer (CC1 mild/standard, Ventana Medical Systems, AZ, USA) using an automated tissue immunostainer (Discovery Staining Module, Ventana Medical Systems, AZ, USA). The staining protocol was set to ‘Mild and Standard Cell Conditioning’ procedure for all antibodies. Primary antibodies were applied for 60 min (except for NF-κBp65 which were applied for 30 min) while secondary antibodies were incubated for 32 min. Initial dilutions quoted above were diluted further 1 : 3 with buffer dispensed onto the slide with the primary antibody. Primary antibodies were omitted for negative controls.
Total ERβ IHC was performed manually; sections were microwaved in the presence of 0.01 M citrate buffer, pH 6.0, for 20 min at full power (Danby, ON, Canada, model DMW 1001 W, 800 W maximum output). Sections were blocked and then incubated using an ERβ monoclonal antibody (14C8, Genetex, TX, USA) at 1 : 100 dilution in a humidified chamber at 4°C overnight, as previously described (Skliris et al, 2002, 2003; Fuqua et al, 2003). Following incubation with biotinylated goat anti-mouse antibody for 60 min at 1 : 200 (Jackson ImmunoResearch Laboratories, PA, USA) and with the Vectastain ABC kit (Vector Laboratories, CA, USA) for 45 min, total ERβ protein was visualised with 3,3′-diaminobenzidine (DAB, Sigma-Aldrich, ON, Canada). Slides were scored semiquantitatively under a standard light microscope. Images were captured using Polaroid DMC-2 software (version 2.0.1, Polaroid, MA, USA).
Quantification technique and marker selection
The expression of ERβ isoforms (full-length-ligand binding ERβ1, total ERβ and ERβ2/cx) and other prognostic markers was assessed using semiquantitative scoring (H-scores). H-scores derive from a semiquantitative assessment of both staining intensity (scale 0–3) and the percentage of positive cells (0–100%), which when multiplied, generates a score ranging from 0 to 300. Tissue microarray staining was evaluated by two authors (GPS, PHW) independently and where discordance was found, cases were re-evaluated together to reach agreement. For the primary categorical analysis, staining and cutoff points to distinguish low from high expression for each marker were as follows: only nuclear staining was evaluated for ERβ1, total ERβ and ERβ2/cx isoforms and since there is no agreement or clinical relevant cutoff IHC-scores for ERβ isoforms reported in the literature, several IHC-score cut-points equivalent to absent staining, the 25th percentile and median IHC-score values were tested in statistical analysis. Ki67, caspase-3 (markers of proliferation and apoptosis, respectively) and CK5/6 (a marker of the basal phenotype) were scored as previously described (Perou et al, 2000; Wykoff et al, 2001; Foulkes et al, 2004; El-Rehim et al, 2005). Since NF-κB has been associated previously with more aggressive breast cancer (Biswas et al, 2004) and both NF-κB and AP-1 have been shown to interact differentially with ERα and ERβ (Paech et al, 1997; An et al, 1999) we have also assessed the relationship of ERβ to these pathways in ERα-negative tumours. For NF-κB/p65 nuclear staining was assessed and multiple H-score cutoffs were tested. P-c-Jun, a marker of AP-1 activity, was defined by nuclear staining and an H-score of >0.
Statistical analysis
Associations between ERβ isoforms and other clinical–pathological variables were tested using contingency methods (Fisher's exact test). Correlations were assessed by the Spearman's rank correlation test (r). Mann–Whitney rank sum tests, two-sided were also used to evaluate variables. Survival analyses were perfomed using the log rank test to generate Kaplan–Meier curves. Overall survival was defined as the time from initial surgery to the date of death attributable to breast cancer. Relapse-free survival was defined as the time from initial surgery to the date of clinically documented local or distant disease recurrence or death attributed to breast cancer. GraphPad Prism 4.02 version statistics software (GraphPad, San Diego, CA, USA) was used to perform all analyses.
Results
Validation of ERβ antibodies
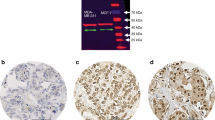
Three antibodies previously validated to detect ERβ related proteins were used in this study (Fuqua et al, 1999; Leav et al, 2001; Saunders et al, 2002). GC17/385P (Leav et al, 2001) was raised to a C-terminal epitope of the wild-type ligand binding isoform of ERβ, generally referred to as ERβ1. 14C8 antibody (Fuqua et al, 1999) was raised to an N-terminal epitope which would be found in both ERβ1 and multiple C-terminal truncated nonligand binding forms of ERβ and therefore would detect multiple known ERβ isoforms including ERβ1 and ERβ2cx. Hence we refer to it as detecting ‘total’ ERβ. The antibody used to detect the nonligand isoform ERβ2/cx (Saunders et al, 2002) has been previously validated by IHC and immunoblotting (Saunders et al, 2002). However, we have also validated the antibody further at the IHC level, by using MCF7 breast cancer cell lines, which have been engineered to overexpress ERβ1 or ERβ2/cx, after induction with the tetracycline analogue doxycycline (Murphy et al, 2005). Agar embedded cell pellets (Riera et al, 1999), formalin-fixed and paraffin-embedded (Adeyinka et al, 2002) from only the doxycycline treated cells expressing ERβ2/cx but not ERβ1 or controls were found to show nuclear staining with the specific ERβ2/cx antibody under the same IHC conditions described above for the human breast tumours (Figure 1A).
(A) Validation of ERβ2/cx antibody (mouse monoclonal, clone 57/3, Serotec, UK): (a) Serotec clone 57/3 antibody staining of section from cell pellet of doxycycline treated tet-on-MDA231 cells stably overexpressing ERβ2/cx, magnification × 500; (b) same as (a), magnification × 1250; (c) Serotec clone 57/3 antibody staining of section from cell pellet of doxycycline treated tet-on-MCF7 cells stably overexpressing ERβ2/cx, magnification × 500; (d) same as (c), magnification × 1250; (e) Serotec clone 57/3 antibody staining of section from cell pellet of a separate clone of doxycycline treated tet-on-MCF7-cells stably overexpressing ERβ2/cx, magnification × 500; (f) same as (e), magnification × 1250; (g) Serotec clone 57/3 antibody staining of section from cell pellet of doxycycline treated tet-on-MCF7 vector alone control cells, magnification × 500; (h) same as (g), magnification × 1250; (i) Serotec clone 57/3 antibody staining of section from cell pellet of doxycycline treated MCF7 stably overexpressing ERβ1 (Murphy et al, 2005), magnification × 500. (B) Expression of ERβcx/2 in ERα-negative invasive tumours and normal breast tissue detected by IHC is demonstrated in representative panels. (a) Tumour core stained with the specific ERβcx/2 antibody (high H-score, 270); (b) tumour stained for ERβcx/2 (low H-score, 25); (c) tumour core showing negative staining for ERβcx/2 H-score, 0); (d) normal breast tissue showing strong, nuclear ERβcx/2 protein expression; (e) nuclear ERβcx/2 expression in normal breast ducts; (f) negative control (omission of ERβcx/2 antibody). Magnification × 500 for a, b, c, and × 1250 for d, e, f.
ERβ isoform expression in ERα-negative human breast tumours
Serial sections of the ERα–255TMA were stained with specific antibodies for ERβ1, total ERβ, and ERβ2/cx using IHC. Nuclear staining could be observed with ERβ1 and total ERβ antibodies in epithelial cells in our series of invasive cancers (Figure 2). Strong nuclear staining in both normal and neoplastic breast tissues for ERβ2/cx isoform was often observed (Figure 1B). Using the 25% percentile of IHC-scores to define positive status for ERβ1, total ERβ and ERβ2/cx, we observed that 58% of ERα-negative tumours were positive for ERβ1 (n=122/210), 60% positive for total ERβ (n=115/192) and 57% of cancers were positive for ERβ2/cx (n=114/199; Table 2).
Expression of ERβ and Ki67 in ERα-negative tissue microarray cores. (A–C) ERα-negative tumour cores stained with the specific ERβ1 antibody (GC17/385P) showing negative, medium and high expression (a–c; H-scores of 0, 150 and 225, respectively); (D–F) ERα-negative tumour cores stained with total ERβ antibody (14C8) showing negative, low and high expression (H-scores of 0, 25 and 100, respectively); (G–I) ERα-negative tumour cores showing negative, medium and high expression for Ki67, a proliferation marker (% positive, 0, 60 and 90%, respectively). Magnification × 500.
ERβ1 was significantly correlated with both total ERβ and ERβ2/cx (r=0.28, P<0.0001, n=189; r=0.27, P=0.0002, n=196, respectively; Table 3). The same relationship was evident in categorical analysis using a variety of cutoff values for contingency analysis, where ERβ1 was also significantly associated with ERβ2/cx and total ERβ (P=0.0083, >10; P=0.0016, 0.0391; >10, >25 respectively, Fishers exact test). Using a cut-point for ERβ1 of either >10 or >25, median levels of total ERβ expression were significantly higher in ERβ1-positive vs -negative tumours (P=0.0026 and P=0.011, Mann–Whitney rank sum tests, two-sided). Similarly using the same two cut-points for ERβ1 positivity median levels of ERβ2/cx expression were significantly higher in ERβ1-positive vs -negative tumours (P=0.0024 and P=0.022, respectively Mann–Whitney rank sum tests). These data suggest frequent coexpression of multiple ERβ isoforms in breast tumours.
Relationship of ERβ isoform expression with markers of proliferation and apoptosis in ERα-negative human breast tumours
ERβ1 (r=0.28, P<0.0001, n=209) and total ERβ (r=0.29, P<0.0001, n=191; Table 3) were positively correlated with Ki67, a marker of proliferation, which was detected in the nuclei of ERα-negative tumours (Figure 2). Contingency analyses also showed that ERβ1 and total ERβ were associated with Ki67 (data not shown). Using the median Ki67 IHC-score as a cutoff to define low Ki67 (⩽25) and high Ki67 (>25), the median level of ERβ1 expression was significantly lower in low Ki67 expressors (median ERβ1=25) compared to high Ki67 expressors (median ERβ1=50; P=0.0008, Mann–Whitney rank sum test). Similarly the median level of total ERβ expression was significantly lower in low Ki67 expressors (median total ERβ=20) compared to high Ki67 expressors (median total ERβ=50; P=0.0008, Mann–Whitney rank sum test). No significant differences in ERβ2/cx were found between the low and high Ki67 groups.
However, high proliferation in primary tumours prior to treatment, is often associated with high levels of apoptosis (Lipponen et al, 1994; Lipponen, 1999; Parton et al, 2002). Therefore, ERβ expression was investigated with respect to a marker of apoptosis, active caspase-3 (Parton et al, 2002). No correlations were detected between ERβ isoforms and caspase-3. However, Ki67 expression was significantly correlated (r=0.44, P<0.0001, n=211, Table 3) and associated (P<0.0001 Fisher's exact test; Mann–Whitney rank sum test) with caspase-3 in this breast tumour cohort. These data suggest that ERβ expression in ERα-negative tumours is associated with markers of a high proliferative index.
Relationship of ERβ expression to basal epithelial phenotype markers in ERα-negative human breast tumours
Invasive breast cancers expressing the basal epithelial phenotype, based on the consensus of the published literature from cDNA microarray and IHC analyses, are ERα negative (Perou et al, 2000; Sorlie et al, 2001; Vijver et al, 2002; Nielsen et al, 2004; El-Rehim et al, 2005), CK5/6 positive (Sorlie et al, 2001; Korsching et al, 2002; Nielsen et al, 2004; Collett et al, 2005) and/or CK14 (El-Rehim et al, 2005) positive. The basal phenotype has also been associated with mutated BRCA1 (Foulkes et al, 2003, 2004; Sorlie et al, 2003; Collett et al, 2005). We were therefore interested to determine the relationship of ERβ expression in ERα-negative tumours to markers of the basal epithelial phenotype. ERβ1 and total ERβ expression were weakly correlated with CK5/6 (r=0.20, P=0.010; n=170; r=0.18, P=0.022, n=158; Table 3). No correlations were seen with ERβ2/cx. These data support the conclusion that many ERα-negative tumours expressing ERβ are associated with some markers of a basal epithelial phenotype in breast cancer.
ERβ2/cx expression in ERα-negative human breast tumours
Despite the correlations and associations of ERβ2/cx to ERβ1 and total ERβ described above, ERβ2/cx was not correlated with Ki67 nor activated caspase-3. However, ERβ2/cx was strongly correlated with p-c-Jun IHC-score (r=0.53, P<0.0001, n=93; Table 3). Contingency analyses for ERβ2/cx and p-c-Jun positivity, identified a significant association of ERβ2/cx with p-c-Jun (P<0.0001, Fisher's exact test). When p-c-Jun expression level was examined in relation to ERβ2/cx status, p-c-jun IHC-score was significantly lower in ERβ2/cx-negative tumours (median p-c-Jun=5) compared to high ERβ2/cx expressors (median p-c-Jun=40; P<0.0001, Mann–Whitney rank sum test).
Similarly, ERβ2/cx expression was also correlated with NF-κBp65 (r=0.35, P<0.0001, n=176; Table 3). Using either the 25% percentile (>0) or the median (>25) ERβ2/cx IHC-score as cut-points to define negative and positive ERβ2/cx status the median level of NF-κBp65 expression was significantly lower in negative/low ERβ2/cx expressors (median NF-κBp65=50) compared to high ERβ2/cx expressors (median NF-κBp65=100; P<0.0001, Mann–Whitney rank sum test). Similar but weaker relationships were found for total ERβ. Using the median (>25) total ERβ IHC-score as a cutoff to define negative and positive total ERβ status the median level of NF-κBp65 expression was significantly lower in negative/low total ERβ expressors (median NF-κBp65=75) compared to high total ERβ expressors (median NF-κBp65=100; P<0.026, Mann–Whitney rank sum test). These data suggest that ERβ2/cx expression is associated with AP1 and NF-κB activity in ERα-negative breast tumours. A relationship between total ERβ and p-c-Jun and NF-κBp65 was also demonstrated, and is likely to reflect the influence of the ERβ2/cx component of the total ERβ signal.
ERβ isoform expression in relation to clinical and pathological prognostic variables and survival
Only total ERβ was associated with tumour grade (P=0.03). No other statistically significant associations between ER isoforms and established prognostic variables such as tumour size, age at diagnosis, node status, inflammation or progesterone receptor, were observed (Table 2, showing associations with cut-points equivalent to the 25th percentile).
Univariate survival analyses in relation to axillary nodal status, size, grade, Ki67, active caspase-3, or basal phenotype, showed a significant association only with nodal status (P=0.024) in this cohort of ERα-negative tumours. Furthermore no difference in disease outcome (overall survival and relapse-free survival) was found between low and high ERβ1, total ERβ or ERβ2/cx (Figure 3).
Kaplan–Meier graphs for ‘overall survival’ and ‘relapse-free survival-time to progression’ with respect to expression of ERβ1 (A and B), ERβ2cx (C and D) and total ERβ isoforms (E and F, respectively). ERβ1 overall survival (A), n=210, low ERβ1 events=47, high ERβ1 events=60. ERβ1 time to progression (B), low ERβ1 events=48, high ERβ1 events=60. ERβ2cx overall survival (C), n=199, low ERβ2cx events=44, high ERβ2cx events=53. ERβ2cx time to progression (D), low ERβ2cx events=44, high ERβ2cx events=53. Total ERβ overall survival (E), n=192, low total ERβ events=40, high total ERβ events=55. Total ERβ time to progression, low total ERβ events=40, high total ERβ events=56.
Discussion
Several interesting observations have been made in the present study concerning ERβ isoform expression in ERα-negative breast tumours. The first is that ERβ1, total ERβ and ERβ2/cx isoforms are frequently expressed in this cohort of ERα-negative breast cancers. The second is that there is a significant correlation of ERβ1 and total ERβ with Ki67, a marker of proliferation, which is of particular interest. As this was not found when ERβ2/cx expression was assessed, it is likely that the correlation with total ERβ reflects the ERβ1 component, although we cannot exclude the existence of other, as yet unknown variant isoforms. Indeed, the frequent expression of the ERβ variant isoform, ERβ5, in ERα-negative breast tumours has recently been described (Poola et al, 2005), however, we did not have access to specific antibodies to investigate this variant isoform in our breast tumour cohort. Our data confirm and extend an observation made by Jensen et al (2001), where the highest expression of either Ki67 and Cyclin A was found in tumours that only expressed ERβ, indicating that ERβ may be related to proliferation in breast cancer. Jensen's observation showing an association of ERβ with Ki67, using an antibody that recognised total ERβ (Jensen et al, 2001), also suggests that ERβ isoforms are not only expressed in cells with the potential to cycle but also can be expressed in cells that are cycling. The existence of this relationship was reflected only in a very small subset of seven tumours in the ERα-negative/ERβ-positive cohort in his study (Jensen et al, 2001), but a study by O'Neill et al (2004) published during the execution of our study confirmed his observation in a larger cohort (n=167). However, results from these latter studies came only from subset analysis of mixed cohorts of ERα-positive and -negative tumours. Our study is the only one so far exclusively focusing on ERα-negative cancers to address the issue of ERβ expression. The cohort used in our study (n=216) is the largest so far and included tumours that were all selected to be ERα negative, both immunohistochemically and by LBA. Thus, the relationship of ERβ1 alone expression in human breast cancer to Ki67, seems to be highly reproducible and therefore likely offers a new significant insight into the possible role of ERβ1 in breast cancer. In contrast, this relationship is generally not seen in ERα-positive/ERβ-positive breast tumours (Jarvinen et al, 2000; Mann et al, 2001; Omoto et al, 2001; Murphy et al, 2002; Fuqua et al, 2003; Iwase et al, 2003; Fleming et al, 2004; Hopp et al, 2004; Myers et al, 2004; Nakopoulou et al, 2004) and therefore our data together with two other studies support the conclusion that the role of ERβ1 when expressed alone in human breast cancers in vivo is likely quite different to when it is coexpressed with ERα. Such data suggest that ERβ1 may have a direct role in proliferation in ERα-negative breast cancers, but this is unproven.
The involvement of ERβ isoforms in proliferation using cell line models is unclear. Most cell line models in which ERβ1 has been stably expressed either inducibly or constitutively show that overexpression of ERβ1 inhibits proliferation irrespective of whether it is coexpressed with ERα (Paruthiyil et al, 2004; Strom et al, 2004; Murphy et al, 2005) or not (Lazennec et al, 2001; Cheng et al, 2004). However, two studies using cell line models have been published in which stable constitutive overexpression of ERβ1 resulted in increased proliferation (Tonetti et al, 2003; Hou et al, 2004) although in the former publication the short form of ERβ1 (truncated by 45 amino acids from the N-terminus) was used. Both breast cancer cell lines used (MDA-MB-231 and MDA-MB-435) are typically ERα negative and therefore can be considered to represent the ERβ alone expressing breast tumours cohort in vivo. However, in another constitutive ERβ overexpression model based on the MDA-MB-231 cells, little or no effect on proliferation, positive or negative, was seen (Rousseau et al, 2004). Such data indicate that differences in potential cell line background, the type of ERβ isoforms expressed and experimental variables including possibly clonal selection can influence the effect of ERβ on proliferation. However, in other cancer cells types where ERβ1 has been overexpressed, increased ERβ1 is most often associated with inhibition of proliferation and/or increased apoptosis (Qiu et al, 2002; Cheng et al, 2004). It is unclear, however, whether the overexpression of ERβ1 in experimental cancer cell line models, is relevant to the levels of ERβ1 seen in tumours in vivo, especially since generally ERβ1 expression is reduced in tumours compared to normal tissues in multiple cancers (Foley et al, 2000; Roger et al, 2001; Skliris et al, 2003) leading to the suggestion that ERβ1 is a tumour-suppressor gene, and certainly would be consistent with the hypothesis that it is antiproliferative (Weihua et al, 2000; Forster et al, 2002; Paruthiyil et al, 2004). As well the possibility exists that ERβ1 may be frequently mutated and/or altered post-translationally in breast cancers in vivo, although no published data as yet address this issue to our knowledge.
ERβ1 and total ERβ isoforms were also significantly correlated with CK5/6, a marker of the basal epithelial phenotype as defined from DNA microarray and IHC analyses, predominantly as ERα negative and CK5/6 positive (Sorlie et al, 2001; Korsching et al, 2002; Collett et al, 2005; El-Rehim et al, 2005). As ERβ is found widely expressed in the basal myoepithelium (Murphy et al, 2002; Speirs et al, 2002) as well as in luminal epithelial cells in normal human breast tissues, it is possible that many ER-negative breast cancers expressing ERβ are derived from a myoepithelial cell lineage, and that ERβ is a marker of this lineage. Interestingly, a reduced myoepithelial cell layer is found in the lactating mammary gland of the ERβ knockout mouse in contrast to the wild-type controls (Forster et al, 2002). This led to the hypothesis that ERβ may be involved in regulating pathways, which are required for the differentiation of the myoepithelial cell lineage in the mammary gland (Forster et al, 2002).
While proliferation and the basal phenotype have been associated with poor survival, no differences in clinical outcome were identified between high and low Ki67 or any markers of the basal phenotype in our ERα-negative breast cancer cohort. It is possible that the lack of association of any of parameters investigated here (ERβ isoforms, Ki67 and caspase-3) with clinical outcome (disease-free survival and overall survival) is confounded by the variety of treatments the patient cohort later received. In addition, most other studies where Ki67 has been examined as a prognostic factor have included both ER-positive and ER-negative tumours in their cohorts (Trihia et al, 2003). It should also be noted that ERα-negative status in our cohort was defined by negative IHC and ligand binding assay. This definition eliminated 15% of an initial ERα-negative series selected only on the basis of ligand binding assay. A similar number of ERα IHC-negative tumours have been found to be positive by ligand binding assay (Huang et al, 1997). The basis for discrepancy between these two ERα assays has been a subject of past discussion in the literature (Huang et al, 1997), but is likely to reflect biological variables rather than tissue selection or composition, because of the design of our tumour bank. Therefore, the current study used stringently defined ERα-negative tumours and so was enriched for a generally more aggressive group of breast tumours.
In comparison to ERβ1, the role of its variant, ERβ2/cx, is even more unclear. Transient expression studies using human ERβ2/cx, have shown that human ERβ2/cx is unable to bind ligand and when overexpressed sufficiently can inhibit ERα transcriptional activity (Ogawa et al, 1998; Peng et al, 2003) but has little if any effect on ERβ1 activity. In breast cancer ERβ2/cx has been identified at both the RNA and protein levels (Saji et al, 2002; Esslimani-Sahla et al, 2004), and now with another antibody we have also shown the presence of ERβ2/cx in both normal and neoplastic breast tissue. Most studies previously published suggested that ERβ2/cx is increased in breast tumours compared to normal breast tissue (Omoto et al, 2002; Palmieri et al, 2004) and the relative expression of the ERβ2/cx to ERβ1 is likely to change during breast tumourigenesis. However, no studies focusing only on ERα-negative tumours have been published. A few studies have suggested hypotheses as to ERβ2/cx function due to observed correlations and association with other prognostic markers and clinical outcome with or without treatment (Omoto et al, 2002; Esslimani-Sahla et al, 2004; Palmieri et al, 2004). Esslimani-Sahla et al (2004) showed that ERβ2/cx expression was correlated with total ERβ, which is in agreement with our observation in our ERα-negative series. However, among these studies contradictory conclusions have often been reached (Saji et al, 2002; Esslimani-Sahla et al, 2004; Palmieri et al, 2004). Our data suggest that in ERα-negative tumours, ERβ2/cx expression is significantly associated with both increased AP-1 and NF-κB expression and that ERβ1 may not be associated with these activities. This suggests that the different ERβ isoforms may be involved in regulation of distinct pathways in these tumours or alternatively there is differential regulation of ERβ isoforms by distinct pathways in these tumours.
The absence of any significant correlations between ERβ isoforms and particularly total ERβ with either overall or relapse-free survival is also in agreement with some other published studies (Hopp et al, 2004) but disagrees with other studies where increased ERβ has been associated with better survival (Nakopoulou et al, 2004) and when patients were treated with tamoxifen alone, where an association was shown with better response to tamoxifen therapy (Murphy and Watson, 2006). However, in these latter studies the majority if not all the tumours assessed were ERα positive and so represent a different context where ERβ is coexpressed with ERα. In the current study we have hypothesised that the function of ERβ expressed alone will be different to that when ERβ is coexpressed with ERα, and therefore we have looked at a distinct cohort of patients where their tumours are ERα negative.
These data support the hypothesis that the role of ERβ expression is different when expressed alone, to its role when coexpressed with ERα in human breast cancer. This is specifically reflected in the present study, by the confirmation of a strong relationship of ERβ1 with Ki67 in ERα-negative tumours, such that it seems likely that the addition of an ERβ1 antagonist could be a potentially useful therapy in specific subsets of breast cancer patients in a clinical setting.
Change history
16 November 2011
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Adeyinka A, Niu Y, Cherlet T, Snell L, Watson P, Murphy L (2002) Activated mitogen-activated protein kinase expression during human breast tumorigenesis and breast cancer progression. Clin Cancer Res 8: 1747–1753
An J, Ribeiro R, Webb P, Gustafsson J, Kushner P, Baxter J, Leitman D (1999) Estradiol repression of tumor necrosis factor-alpha transcription requires estrogen receptor activation function-2 and is enhanced by coactivators. Proc Natl Acad Sci USA 96: 15161–15166
Biswas D, Shi Q, Baily S, Strickland I, Ghosh S, Pardee A, Iglehart J (2004) NF-kappa B activation in human breast cancer specimens and its role in cell proliferation and apoptosis. Proc Natl Acad Sci USA 101: 10137–10142
Brouillet J, Dujardin M, Chalbos D, Rey J, Grenier J, Lamy P, Maudelonde T, Pujol P (2001) Analysis of the potential contribution of estrogen receptor (ER) b in ER cytosolic assay of breast cancer. Int J Cancer 95: 205–208
Cheng J, Lee E, Madison L, Lazennec G (2004) Expression of estrogen receptor beta in prostate carcinoma cells inhibits invasion and proliferation and triggers apoptosis. FEBS Lett 566: 169–172
Collett K, Stefansson I, Eide J, Braaten A, Wang H, Eide G, SOThoresen Foulkes W, Akslen L (2005) A Basal epithelial phenotype is more frequent in interval breast cancers compared with screen detected tumors. Cancer Epidemiol Biomarkers Prev 14: 1108–1112
El-Rehim DA, Ball G, Pinder S, Rakha E, Paish C, Robertson J, Macmillan D, Blamey R, Ellis I (2005) High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int J Cancer 116: 340–350
Esslimani-Sahla M, Simony-Lafontaine J, Kramar A, Lavaill R, Mollevi C, Warner M, Gustafsson J-A, Rochefort H (2004) Estrogen receptor beta (ERbeta) level but not its ERbeta-cx variant helps to predict tamoxifen resistance in breast cancer. Clin Cancer Res 10: 5769–5776
Fleming F, Hill A, McDermott E, O'Higgins N, Young L (2004) Differential recruitment of coregulator proteins steroid receptor coactivator-1 and silencing mediator for retinoid and thyroid receptors to the estrogen receptor-estrogen response element by beta-estradiol and 4-hydroxytamoxifen in human breast cancer. J Clin Endocrinol Metab 89: 375–383
Foley E, Jazaeri A, Shupnik M, Jazaeri O, Rice L (2000) Selective loss of estrogen receptor beta in malignant human colon. Cancer Res 60: 245–248
Forster C, Makela S, Warri A, Kietz S, Becker D, Hultenby K, Warner M, Gustafsson J (2002) Involvement of estrogen receptor b in terminal differentiation of mammary gland epithelium. Proc Natl Acad Sci USA 99: 15578–15583
Foulkes W, Brunet J, Stefansson I, Straume O, Chappuis P, Begin L, Hamel N, Goffin J, Wong N, Trudel M, Kapusta L, Porter P, Akslen L (2004) The prognostic implication of the basal-like (cyclin E high/p27 low/p53+/glomeruloid-microvascular-proliferation+) phenotype of BRCA1-related breast cancer. Cancer Res 64: 830–835
Foulkes W, Stefansson I, Chappuis P, Begin L, Goffin J, Wong N, Trude M, Akslen L (2003) Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst 95: 1482–1485
Fuqua S, Schiff R, Parra I, Friedrichs W, Su J, McKee D, Slentz-Kesler K, Moore L, Willson T, Moore J (1999) Expression of wild-type estrogen receptor beta and variant isoforms in human breast cancer. Cancer Res 59: 5425–5428
Fuqua S, Schiff R, Parra I, Moore J, Mohsin S, Osborne C, Clark G, Allred D (2003) Estrogen receptor beta protein in human breast cancer: correlation with clinical tumor parameters. Cancer Res 63: 2434–2439
Harvey J, Clark G, Osborne C, Allred D (1999) Estrogen receptor status by immunohistochemistry is superior to the ligand binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol 17: 1474–1481
Hopp T, Weiss H, Parra I, Cui Y, Osborne C, Fuqua S (2004) Low levels of estrogen receptor beta protein predict resistance to tamoxifen therapy in breast cancer. Clin Cancer Res 10: 7490–7499
Hou Y, Yuan S, Li H, Wu J, Lu J, Liu G, Lu L, Shen Z, Ding J, Shao Z (2004) ERbeta exerts multiple stimulative effects on human breast carcinoma cells. Oncogene 23: 5799–5806
Huang A, Leygue E, Snell L, Murphy L, Watson P (1997) Expression of estrogen receptor variants mRNAs and determination of estrogen status in human breast cancer. Am J Pathol 150: 1827–1833
Iwase H, Zhang Z, Omoto Y, Sugiura H, Yamashita H, Toyama T, Iwata H, Kobayashi S (2003) Clinical significance of the expression of estrogen receptors alpha and beta for endocrine therapy of breast cancer. Cancer Chemother Pharmacol 52 (Suppl 1): S34–S38
Jarvinen T, Pelto-Huikko M, Holli K, Isola J (2000) Estrogen receptor beta is coexpressed with ERalpha and PR and associated with nodal status, grade, and proliferation rate in breast cancer. Am J Pathol 156: 29–35
Jensen E, Cheng G, Palmieri C, Saji S, Makela S, Noorden Sv, Wahlstrom T, Warner M, Coombes R, Gustafsson J-A (2001) Estrogen receptors and proliferation markers in primary and recurrent breast cancer. Proc Natl Acad Sci USA 98: 15197–15202
Korsching E, Packeisen J, Agelopoulos K, Eisenacher M, Voss R, Isola J, Diest Pv, Brandt B, Boecker W, Buerger H (2002) Cytogenetic alterations and cytokeratin expression patterns in breast cancer: integrating a new model of breast differentiation into cytogenetic pathways of breast carcinogenesis. Lab Invest 82: 1525–1533
Lapidus R, Nass S, Davidson N (1998) The loss of estrogen and progesterone receptor gene expression in human breast cancer. J Mammary Gland Biol Neoplasia 3: 85–94
Lazennec G, Bresson D, Lucas A, Chauveau C, Vignon F (2001) ERb inhibits proliferation and invasion of breast cancer cells. Endocrinol 142: 4120–4130
Leav I, Lau K-M, Adams J, McNeal J, Taplin M-E, Wang J, Singh H, Ho S-M (2001) Comparative studies of the estrogen receptor b and a and the androgen receptor in normal human prostate glands, dysplasia and in primary and metastatic carcinoma. Am J Pathol 159: 79–92
Leygue E, Dotzlaw H, Watson P, Murphy L (1998) Altered estrogen receptor alpha and beta mRNA expression during human breast tumorigenesis. Cancer Res 58: 3197–3201
Leygue E, Dotzlaw H, Watson P, Murphy L (1999) Expression of estrogen receptor beta1, beta2, and beta5 messenger RNAs in human breast tissue. Cancer Res 59: 1175–1179
Lipponen P (1999) Apoptosis in breast cancer: relationship with other pathological parameters. Endocr Relat Cancer 6: 13–16
Lipponen P, Aaltomaa S, Kosma V, Syrjanen K (1994) Apoptosis in breast cancer as related to histopathological characteristics and prognosis. Eur J Cancer 30A: 2068–2073
Lu B, Leygue E, Dotzlaw H, Murphy L, Murphy L, Watson P (1998) Estrogen receptor-beta mRNA variants in human and murine tissues. Mol Cell Endocrinol 138: 199–203
Mann S, Laucirica R, Carlson N, Younes P, Ali N, Younes A, Li Y, Younes M (2001) estrogen receptor beta expression in invasive breast cancer. Hum Pathol 32: 113–118
Murphy L, Cherlet T, Lewis A, Banu Y, Watson P (2003) New insights into estrogen receptor function in human breast cancer. Ann Med 35: 614–631
Murphy L, Leygue E, Niu Y, Snell L, Ho S-M, Watson P (2002) Relationship of coregulator and estrogen receptor isoform expression to de novo tamoxifen resistance in human breast cancer. Br J Cancer 87: 1411–1416
Murphy L, Peng B, Lewis A, Davie J, Leygue E, Kemp A, Ung K, Vendetti M, Shiu R (2005) Inducible upregulation of oestrogen receptor-beta1 affects oestrogen and tamoxifen responsiveness in MCF7 human breast cancer cells. J Mol Endocrinol 34: 553–566
Murphy L, Watson P (2006) Is Oestrogen Receptor-beta a Predictor of Endocrine Therapy Responsiveness in Human Breast Cancer? Endocr Relat Cancer 13: 327–334
Myers E, Fleming F, Crotty T, Kelly G, McDermott E, O'Higgins N, Hill A, Young L (2004) Inverse relationship between ER-beta and SRC-1 predicts outcome in endocrine resistant breast cancer. Br J Cancer 91: 1687–1693
Nakopoulou L, Panayotopoulou AL, Giannopoulou I, Givalos N, Markaki S, Keramopoulos A (2004) The favourable prognostic value of oestrogen receptor beta immunohistochemical expression in breast cancer. J Clin Pathol 57: 523–528
Nielsen T, Hsu R, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L, Akslen L, Ragaz J, Gown A, Gilks C, Rijn MVD, Perou C (2004) Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res 10: 5367–5374
O'Neill P, Davies M, Shaaban A, Innes H, Torevell A, Sibson D, Foster C (2004) Wild-type oestrogen receptor beta (ERbeta1) mRNA and protein expression in Tamoxifen-treated post-menopausal breast cancers. Br J Cancer 91: 1694–1702
Ogawa S, Inoue S, Watanabe T, Orimo A, Hosoi T, Ouchi Y, Muramatsu M (1998) Molecular cloning and characterization of human estrogen receptor bcx: potential inhibitor of estrogen action in human. Nucleic Acids Res 26: 3505–3512
Omoto Y, Inoue S, Ogawa S, Toyama T, Yamashita H, Muramatsu M, Kobayashi S, Iwase H (2001) Clinical value of the wild-type estrogen receptor beta expression in breast cancer. Cancer Lett 163: 207–212
Omoto Y, Kobayashi S, S Inoue S, Ogawa S, Toyama T, Yamashita H, Muramatsu M, Gustafsson J, Iwase H (2002) Evaluation of oestrogen receptor beta wild-type and variant protein expression, and relationship with clinicopathological factors in breast cancers. Eur J Cancer 38: 380–386
Osborne C (1998) Steroid hormone receptors in breast cancer management. Breast Cancer Res Treat 51: 227–238
Paech K, Webb P, Kuiper G, Nilsson S, Gustafsson J, angst Kushner PJ, Scanlan TS (1997) Differential ligand activation of estrogen receptors ERalpha and ERbeta at AP1 Sites. Science 277: 1508–1510
Palmieri C, Lam E, Mansi J, MacDonald C, Shousha S, Madden P, Omoto Y, Sunters A, Warner M, Gustafsson J, Coombes R (2004) The expression of ER beta cx in human breast cancer and the relationship to endocrine therapy and survival. Clin Cancer Res 10: 2421–2428
Parton M, Krajewski S, Smith I, Krajewska M, Archer C, Naito M, Ahern R, Reed J, Dowsett M (2002) Coordinate expression of apoptosis-associated proteins in human breast cancer before and during chemotherapy. Clin Cancer Res 8: 2100–2108
Paruthiyil S, Parmar H, Kerekatte V, Cunha G, Firestone G, Leitman D (2004) Estrogen receptor β inhibits human breast cancer cell proliferation and tumor formation by causing a G2 cell cycle arrest. Cancer Res 64: 423–428
Peng B, Lu B, Leygue E, Murphy L (2003) Putative functional characteristics of human estrogen receptor-beta isoforms. J Mol Endocrinol 30: 13–29
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D (2000) Molecular portraits of human breast tumors. Nature 406: 747–752
Poola I, Fuqua S, Witty RD, Abraham J, Marshallack J, Liu A (2005) Estrogen receptor-alpha-negative breast cancer tissues express significant levels of estrogen-independent transcription factors, ER-beta-1 and ER-beta-5: potential molecular targets for chemoprevention. Clin Cancer Res 11: 7579–7585
Qiu Y, Waters C, Lewis A, Langmann M, Eggo M (2002) Oestrogen induced apoptosis in colonocytes expressing oestrogen receptor beta. J Endocrinol 174: 369–377
Riera J, Simpson J, Tamayo R, Battifora H (1999) Use of cultured cells as a control for quantitative immunohistochemical analysis of estrogen receptor in breast cancer. Am J Clin Pathol 111: 329–335
Roger P, Sahla M, Makela S, Gustafsson JA, Baldet P, Rochefort H (2001) Decreased expression of estrogen receptor beta protein in proliferative preinvasive mammary tumors. Cancer Res 61: 2537–2541
Rousseau C, Nichol J, Pettersson F, Couture M, Miller W (2004) ERbeta sensitizes breast cancer cells to retinoic acid: evidence of transcriptional crosstalk. Mol Cancer Res 2: 523–531
Saji S, Omoto Y, Shimizu C, Warner M, Hayashi Y, Horiguchi S, Watanabe T, Hayashi S, Gustafsson J (2002) Expression of estrogen receptor (ER) (beta)cx protein in ER (alpha)-positive breast cancer: specific correlation with progesterone receptor. Cancer Res 62: 4849–4853
Saunders P, Millar M, Macpherson S, Irvine D, Groome N, Evans L, Sharpe R, Scobie G (2002) ERbeta1 and the ERbeta2 splice variant (ERbetacx/beta2) are expressed in distinct cell populations in the adult human testis. J Clin Endocrinol Metab 87: 2706–2715
Skliris G, Munot K, Bell S, Carder P, lane S, Horgan K, Lansdown M, Parkes A, Hanby A, Markham A, Speirs V (2003) Reduced expression of oestrogen receptor beta in invasive breast cancer and its re-expression using DNA methyltransferase inhibitors in a cell line model. J Pathol 201: 213–220
Skliris G, Parkes A, Limer J, Burdall S, Carder P, Speirs V (2002) Evaluation of seven oestrogen receptor β antibodies for immunohistochemistry, western blotting and flow cytometry in human breast tissue. J Pathol 197: 155–162
Sorlie T, Perou C, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen M, Rijn MVd, Jeffrey S, Thorsen T, Quist H, Matese J, Brown P, Botstein D, Lonning P, Borresen-Dale A (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98: 10869–10874
Sorlie T, Tibshirani R, Parker J, Hastie T, Marron J, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou C, Lonning P, Brown P, Borresen-Dale A, Botstein D (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 100: 8418–8423
Speirs V, Skliris G, Burdall S, Carder P (2002) Distinct expression patterns of ERa and ERb in normal human mammary gland. J Clin Pathol 255: 371–374
Strom A, Hartman J, Foster J, Kietz S, Wimalasena J, Gustafsson J-A (2004) Estrogen receptor β inhibits 17b-estradiol-stimulated proliferation of the breast cancer cell line T-47D. Proc Natl Acad Sci USA 101: 1566–1571
Tonetti D, Rubenstein R, Deleon M, Zhao H, Pappas S, Bentrem D, Chen B, Constantinou A, Jordan V (2003) Stable transfection of an estrogen receptor beta cDNA isofrm into MDAMB231 breast cancer cells. J Steroid Biochem Mol Biol 87: 47–55
Trihia H, Murray S, Price K, Gelber R, Golouh R, Goldhirsch A, Coates A, Collins J, Castiglione-Gertsch M, Gusterson B, Group IBCS (2003) Ki-67 expression in breast carcinoma: its association with grading systems, clinical parameters, and other prognostic factors – a surrogate marker? Cancer 97: 1321–1331
Vijver MV, He Y, Veer Lv, Dai H, Hart A, Voskuil D, Schreiber G, Peterse J, Roberts C, Marton M, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, Velde TVD, Bartelink H, Rodenhuis S, Rutgers E, Friend S, Bernards R (2002) A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347: 1999–2009
Watson P, Snell L, Parisien M (1996) The NCIC-Manitoba Breast Tumor Bank: a resource for applied cancer research. CMAJ 155: 281–283
Weihua Z, Saji S, Makinen S, Cheng G, Jensen E, Warner M, Gustaffson J-A (2000) Estrogen receptor (ER) β, a modulator of ERα in the uterus. Proc Natl Acad Sci 97: 5936–5941
Wykoff C, Beasley N, Watson P, Campo L, Chia S, English R, Pastorek J, Sly W, Ratcliffe P, Harris A (2001) Expression of the hypoxia-inducible and tumor-associated carbonic anhydrases in ductal carcinoma in situ of the breast. Am J Pathol 158: 1011–1019
Acknowledgements
GS was funded by a Postdoctoral Fellowship from the Manitoba Health Research Council (MHRC) and previously from the CancerCare Manitoba Foundation (CCMF). The research is supported by Canadian Institutes of Health Research (CIHR), Canadian Breast Cancer Research Inititative (CBCRI), CCMF and USAMRMC operating grants. We acknowledge the strong support of the CCMF for our facilities at MICB. The authors have no known conflicts of interests either financial or personal between themselves and others that might bias the work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Skliris, G., Leygue, E., Curtis-Snell, L. et al. Expression of oestrogen receptor-β in oestrogen receptor-α negative human breast tumours. Br J Cancer 95, 616–626 (2006). https://doi.org/10.1038/sj.bjc.6603295
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.bjc.6603295
Keywords
This article is cited by
-
Estrogen receptor β2 (ERβ2)-mediated upregulation of hsa_circ_0000732 promotes tumor progression via sponging microRNA-1184 in triple-negative breast cancer (TNBC)
Inflammation Research (2022)
-
Expression of estrogen receptor beta correlates with adverse prognosis in resected pancreatic adenocarcinoma
BMC Cancer (2018)
-
Androgen Receptor and Ki67 Expression and Survival Outcomes in Non-small Cell Lung Cancer
Hormones and Cancer (2018)
-
Estrogen receptor beta impacts hormone-induced alternative mRNA splicing in breast cancer cells
BMC Genomics (2015)
-
Ki-67 evaluation at the hottest spot predicts clinical outcome of patients with hormone receptor-positive/HER2-negative breast cancer treated with adjuvant tamoxifen monotherapy
Breast Cancer (2015)