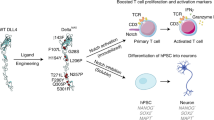
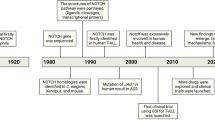
Abstract
Notch receptors transmit signals between adjacent cells. Signaling is initiated when ligand binding induces metalloprotease cleavage of Notch within an extracellular negative regulatory region (NRR). We present here the X-ray structure of the human NOTCH2 NRR, which adopts an autoinhibited conformation. Extensive interdomain interactions within the NRR bury the metalloprotease site, showing that a substantial conformational movement is necessary to expose this site during activation by ligand. Leukemia-associated mutations in NOTCH1 probably release autoinhibition by destabilizing the conserved hydrophobic core of the NRR.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
01 April 2007
References were re-numbered
Notes
*NOTE: In the version of this article initially published, three references were missing from the reference list. The corrected reference appears above and references have been renumbered accordingly. The error has been corrected in the HTML and PDF versions of the article.
References
Bray, S.J. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678–689 (2006).
Weng, A.P. et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306, 269–271 (2004).
Blaumueller, C.M., Qi, H., Zagouras, P. & Artavanis-Tsakonas, S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell 90, 281–291 (1997).
Logeat, F. et al. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. USA 95, 8108–8112 (1998).
Sanchez-Irizarry, C. et al. Notch subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Mol. Cell. Biol. 24, 9265–9273 (2004).
Fehon, R.G. et al. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell 61, 523–534 (1990).
Weinmaster, G. The ins and outs of notch signaling. Mol. Cell. Neurosci. 9, 91–102 (1997).
Brou, C. et al. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol. Cell 5, 207–216 (2000).
Mumm, J.S. et al. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol. Cell 5, 197–206 (2000).
Kopan, R. & Goate, A. A common enzyme connects notch signaling and Alzheimer's disease. Genes Dev. 14, 2799–2806 (2000).
Schroeter, E.H., Kisslinger, J.A. & Kopan, R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393, 382–386 (1998).
Struhl, G. & Adachi, A. Nuclear access and action of notch in vivo. Cell 93, 649–660 (1998).
Struhl, G., Fitzgerald, K. & Greenwald, I. Intrinsic activity of the Lin-12 and Notch intracellular domains in vivo. Cell 74, 331–345 (1993).
Kopan, R., Schroeter, E.H., Weintraub, H. & Nye, J.S. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc. Natl. Acad. Sci. USA 93, 1683–1688 (1996).
Lieber, T., Kidd, S., Alcamo, E., Corbin, V. & Young, M.W. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 7, 1949–1965 (1993).
Rebay, I., Fehon, R.G. & Artavanis-Tsakonas, S. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell 74, 319–329 (1993).
Berry, L.W., Westlund, B. & Schedl, T. Germ-line tumor formation caused by activation of glp-1, a Caenorhabditis elegans member of the Notch family of receptors. Development 124, 925–936 (1997).
Greenwald, I. & Seydoux, G. Analysis of gain-of-function mutations of the lin-12 gene of Caenorhabditis elegans. Nature 346, 197–199 (1990).
Vardar, D., North, C.L., Sanchez-Irizarry, C., Aster, J.C. & Blacklow, S.C. Nuclear magnetic resonance structure of a prototype Lin12-Notch repeat module from human Notch1. Biochemistry 42, 7061–7067 (2003).
Holm, L. & Sander, C. Mapping the protein universe. Science 273, 595–603 (1996).
Maeda, T. et al. Solution structure of the SEA Domain from the murine homologue of ovarian cancer antigen CA125 (MUC16). J. Biol. Chem. 279, 13174–13182 (2004).
Macao, B., Johansson, D.G.A., Hansson, G.C. & Hard, T. Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin. Nat. Struct. Mol. Biol. 13, 71–76 (2006).
Mumm, J.S. & Kopan, R. Notch signaling: from the outside in. Dev. Biol. 228, 151–165 (2000).
Itoh, M. et al. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev. Cell 4, 67–82 (2003).
Le Borgne, R., Bardin, A. & Schweisguth, F. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development 132, 1751–1762 (2005).
Wang, W. & Struhl, G. Drosophila Epsin mediates a select endocytic pathway that DSL ligands must enter to activate Notch. Development 131, 5367–5380 (2004).
Wang, W. & Struhl, G. Distinct roles for Mind bomb, Neuralized and Epsin in mediating DSL endocytosis and signaling in Drosophila. Development 132, 2883–2894 (2005).
Bingham, S. et al. Neurogenic phenotype of mind bomb mutants leads to severe patterning defects in the zebrafish hindbrain. Dev. Dyn. 228, 451–463 (2003).
Lai, E.C., Roegiers, F., Qin, X., Jan, Y.N. & Rubin, G.M. The ubiquitin ligase Drosophila Mind bomb promotes Notch signaling by regulating the localization and activity of Serrate and Delta. Development 132, 2319–2332 (2005).
Le Borgne, R., Remaud, S., Hamel, S. & Schweisguth, F. Two distinct E3 ubiquitin ligases have complementary functions in the regulation of delta and serrate signaling in Drosophila. PLoS Biol. 3, 688–696 (2005).
Le Borgne, R. & Schweisguth, F. Unequal segregation of Neuralized biases Notch activation during asymmetric cell division. Dev. Cell 5, 139–148 (2003).
Pavlopoulos, E. et al. neuralized encodes a peripheral membrane protein involved in delta signaling and endocytosis. Dev. Cell 1, 807–816 (2001).
Seugnet, L., Simpson, P. & Haenlin, M. Requirement for dynamin during Notch signaling in Drosophila neurogenesis. Dev. Biol. 192, 585–598 (1997).
Parks, A.L., Klueg, K.M., Stout, J.R. & Muskavitch, M.A. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development 127, 1373–1385 (2000).
Ahimou, F., Mok, L.P., Bardot, B. & Wesley, C. The adhesion force of Notch with Delta and the rate of Notch signaling. J. Cell Biol. 167, 1217–1229 (2004).
Maskos, K. et al. Crystal structure of the catalytic domain of human tumor necrosis factor-alpha-converting enzyme. Proc. Natl. Acad. Sci. USA 95, 3408–3412 (1998).
Sun, X. & Artavanis-Tsakonas, S. Secreted forms of DELTA and SERRATE define antagonists of Notch signaling in Drosophila. Development 124, 3439–3448 (1997).
Varnum-Finney, B. et al. Immobilization of Notch ligand, Delta-1, is required for induction of notch signaling. J. Cell Sci. 113, 4313–4318 (2000).
Chen, N. & Greenwald, I. The lateral signal for LIN-12/Notch in C. elegans vulval development comprises redundant secreted and transmembrane DSL proteins. Dev. Cell. 6, 183–192 (2004).
Malecki, M.J. et al. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol. Cell. Biol. 26, 4642–4651 (2006).
Otwinowsk, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Perrakis, A., Morris, R. & Lazmin, V. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6, 458–463 (1999).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Brunger, A.T., Adams, P. & Clore, G. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol. Crystallogr. 54, 905–921 (1998).
Murshudov, G., Vagin, A. & Dodson, E. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–245 (1997).
Acknowledgements
We thank Mike Eck and Angela Toms for crystallographic suggestions and Kelly Arnett and Mike Malecki for critical reading of the manuscript. This work was supported by American Cancer Society Postdoctoral Fellowships (WRG and DVU), a Leukemia and Lymphoma Society Fellowship (WRG), and NIH grants to SCB and JCA.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Sequence alignment of the NRR region of various Notch receptors (PDF 2309 kb)
Supplementary Fig. 2
Representative electron density (PDF 370 kb)
Supplementary Fig. 3
Comparison of human NOTCH2 HD domain with SEA domains from mucins (PDF 175 kb)
Rights and permissions
About this article
Cite this article
Gordon, W., Vardar-Ulu, D., Histen, G. et al. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol 14, 295–300 (2007). https://doi.org/10.1038/nsmb1227
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1227
This article is cited by
-
Soluble and multivalent Jag1 DNA origami nanopatterns activate Notch without pulling force
Nature Communications (2024)
-
Affinity-matured DLL4 ligands as broad-spectrum modulators of Notch signaling
Nature Chemical Biology (2023)
-
TM2D3, a mammalian homologue of Drosophila neurogenic gene product Almondex, regulates surface presentation of Notch receptors
Scientific Reports (2023)
-
The role of Hedgehog and Notch signaling pathway in cancer
Molecular Biomedicine (2022)
-
Notch signaling pathway: architecture, disease, and therapeutics
Signal Transduction and Targeted Therapy (2022)