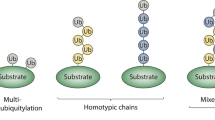
Key Points
-
Ubiquitin is an intracellular signalling molecule that is conjugated to various proteins. Ubiquitin conjugation to itself yields Lys- or Met-conjugated chains, thus expanding its repertoire of signalling networks.
-
Ubiquitin-binding domains (UBDs) are modular elements that bind non-covalently to the protein modifier ubiquitin.
-
Specific ubiquitin–UBD interactions are crucial for the regulation of multiple cellular functions, including protein stability, receptor trafficking, DNA damage responses and inflammatory pathways.
-
UBD preferences for distinct ubiquitin chains of specific length and linkage are mediated through multimeric interactions, sequence context of the UBD and conformational changes following binding.
-
Structures of ubiquitin–UBD complexes have revealed mechanisms of selectivity and specificity in their functional interactions in vivo.
-
Defects in ubiquitin–UBD interactions are relevant for development of disease, such as inflammation and cancer. The new structure-based insights provide strategies for the design of new approaches that can therapeutically target ubiquitin–UBD interaction surfaces.
Abstract
Ubiquitin-binding domains (UBDs) are modular elements that bind non-covalently to the protein modifier ubiquitin. Recent atomic-level resolution structures of ubiquitin–UBD complexes have revealed some of the mechanisms that underlie the versatile functions of ubiquitin in vivo. The preferences of UBDs for ubiquitin chains of specific length and linkage are central to these functions. These preferences originate from multimeric interactions, whereby UBDs synergistically bind multiple ubiquitin molecules, and from contacts with regions that link ubiquitin molecules into a polymer. The sequence context of UBDs and the conformational changes that follow their binding to ubiquitin also contribute to ubiquitin signalling. These new structure-based insights provide strategies for controlling cellular processes by targeting ubiquitin–UBD interfaces.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Pickart, C. M. & Eddins, M. J. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695, 55–72 (2004).
Varshavsky, A. The ubiquitin system. Trends Biochem. Sci. 22, 383–387 (1997).
Ikeda, F. & Dikic, I. Atypical ubiquitin chains: new molecular signals. EMBO Rep. 9, 536–542 (2008).
Kirisako, T. et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 25, 4877–4887 (2006). The description of a complex containing two RING finger proteins that can form linear (Met to Gly) linkages.
Iwai, K. & Tokunaga, F. Linear polyubiquitination: a new regulator of NF-κB activation. EMBO Rep. 10, 706–713 (2009).
Tokunaga, F. et al. Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nature Cell Biol. 11, 123–132 (2009). NEMO is identified as the first in vivo target of the LUBAC ligase complex. Linear ubiquitylation of NEMO is crucial for activation of the NF-kB, but not the Jun N-terminal kinase, pathway in vivo.
Rahighi, S. et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell 136, 1098–1109 (2009). The first structure of a UBD in complex with linear ubiquitin chains. The authors also report that the UBAN domain of the adaptor NEMO is crucial for binding to linear ubiquitin chains and for activation of the NF-kB pathway in vivo.
Lange, O. F. et al. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science 320, 1471–1475 (2008).
Pickart, C. M. & Fushman, D. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 8, 610–616 (2004).
Komander, D. et al. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 10, 466–473 (2009).
Cook, W. J. et al. Structure of tetraubiquitin shows how multiubiquitin chains can be formed. J. Mol. Biol. 236, 601–609 (1994).
Eddins, M. J. et al. Mms2–Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nature Struct. Mol. Biol. 13, 915–920 (2006).
Ryabov, Y. & Fushman, D. Interdomain mobility in di-ubiquitin revealed by NMR. Proteins 63, 787–796 (2006).
Sato, Y. et al. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature 455, 358–362 (2008).
Newton, K. et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 134, 668–678 (2008).
Di Fiore, P. P. et al. When ubiquitin meets ubiquitin receptors: a signalling connection. Nature Rev. Mol. Cell Biol. 4, 491–497 (2003).
Hicke, L. et al. Ubiquitin-binding domains. Nature Rev. Mol. Cell Biol. 6, 610–621 (2005).
Hofmann, K. Ubiquitin-binding domains and their role in the DNA damage response. DNA Repair (Amst.) 8, 544–556 (2009).
Hurley, J. H. et al. Ubiquitin-binding domains. Biochem. J. 399, 361–372 (2006).
Harper, J. W. & Schulman, B. A. Structural complexity in ubiquitin recognition. Cell 124, 1133–1136 (2006).
Bomar, M. G. et al. Structure of the ubiquitin-binding zinc finger domain of human DNA Y-polymerase ɛ. EMBO Rep. 8, 247–251 (2007).
Hirano, S. et al. Double-sided ubiquitin binding of Hrs-UIM in endosomal protein sorting. Nature Struct. Mol. Biol. 13, 272–277 (2006).
Lee, S. et al. Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5. Nature Struct. Mol. Biol. 13, 264–271 (2006).
Penengo, L. et al. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell 124, 1183–1195 (2006). References 24 and 25 reveal a new binding surface on ubiquitin for the ZnF UBDs of RABEX5.
Swanson, K. A. et al. Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor downregulation. EMBO J. 22, 4597–4606 (2003).
Wang, Q. et al. Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. J. Mol. Biol. 348, 727–739 (2005).
Chang, Y. G. et al. Solution structure of the ubiquitin-associated domain of human BMSC-UbP and its complex with ubiquitin. Protein Sci. 15, 1248–1259 (2006).
Kang, R. S. et al. Solution structure of a CUE-ubiquitin complex reveals a conserved mode of ubiquitin binding. Cell 113, 621–630 (2003).
Ohno, A. et al. Structure of the UBA domain of Dsk2p in complex with ubiquitin molecular determinants for ubiquitin recognition. Structure 13, 521–532 (2005).
Swanson, K. A. et al. Structural basis for monoubiquitin recognition by the Ede1 UBA domain. J. Mol. Biol. 358, 713–724 (2006).
Brzovic, P. S. et al. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol. Cell 21, 873–880 (2006).
Hirano, S. et al. Structural basis of ubiquitin recognition by mammalian Eap45 GLUE domain. Nature Struct. Mol. Biol. 13, 1031–1032 (2006).
Alam, S. L. et al. Structural basis for ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain. Nature Struct. Mol. Biol. 13, 1029–1030 (2006).
Schreiner, P. et al. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 453, 548–552 (2008).
Husnjak, K. et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453, 481–488 (2008). References 35 and 36 describe the structure and function of Rpn13 as a new proteasomal receptor for ubiquitin. The PRU domain of Rpn13 binds ubiquitin through loops rather than through secondary structural elements.
Alam, S. L. et al. Ubiquitin interactions of NZF zinc fingers. EMBO J. 23, 1411–1421 (2004).
Bienko, M. et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310, 1821–1824 (2005).
Reyes-Turcu, F. E. et al. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell 124, 1197–1208 (2006). Reveals a unique binding mode between ZnF UBDs and the C-terminal diGly motif of ubiquitin chains.
Amerik, A. et al. In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. EMBO J. 16, 4826–4838 (1997).
Komander, D. et al. Breaking the chains: structure and function of the deubiquitinases. Nature Rev. Mol. Cell Biol. 10, 550–563 (2009).
Raasi, S. et al. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nature Struct. Mol. Biol. 12, 708–714 (2005).
Varadan, R. et al. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol. Cell 18, 687–698 (2005).
Komander, D. et al. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol. Cell 29, 451–464 (2008).
Kim, H. T. et al. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 282, 17375–17386 (2007).
Sobhian, B. et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316, 1198–1202 (2007).
Wang, B. et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316, 1194–1198 (2007).
Sims, J. J. & Cohen, R. E. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol. Cell 33, 775–783 (2009). Shows that specificity for Lys63- or Lys48-linked chains can be determined by the sequence between the two UBDs, as is the case for RAP80 and ataxin 3.
Chai, Y. et al. Poly-ubiquitin binding by the polyglutamine disease protein ataxin-3 links its normal function to protein surveillance pathways. J. Biol. Chem. 279, 3605–3611 (2004).
Wagner, S. et al. Ubiquitin binding mediates the NF-κB inhibitory potential of ABIN proteins. Oncogene 27, 3739–3745 (2008).
Lo, Y. C. et al. Structural basis for recognition of diubiquitins by NEMO. Mol. Cell 33, 602–615 (2009).
Chiu, Y. H. et al. Ubiquitin in NF-κB signaling. Chem. Rev. 109, 1549–1560 (2009).
Oshima, S. et al. ABIN-1 is a ubiquitin sensor that restricts cell death and sustains embryonic development. Nature 457, 906–909 (2009).
Haglund, K. et al. Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem. Sci. 28, 598–603 (2003).
Raiborg, C. & Stenmark, H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458, 445–452 (2009).
Saksena, S. et al. ESCRTing proteins in the endocytic pathway. Trends Biochem. Sci. 32, 561–573 (2007).
Williams, R. L. & Urbe, S. The emerging shape of the ESCRT machinery. Nature Rev. Mol. Cell Biol. 8, 355–368 (2007).
Prag, G. et al. Structural mechanism for ubiquitinated-cargo recognition by the Golgi-localized, γ-ear-containing, ADP-ribosylation-factor-binding proteins. Proc. Natl Acad. Sci. USA 102, 2334–2339 (2005).
Puertollano, R. & Bonifacino, J. S. Interactions of GGA3 with the ubiquitin sorting machinery. Nature Cell Biol. 6, 244–251 (2004).
Akutsu, M. et al. Structural basis for recognition of ubiquitinated cargo by Tom1-GAT domain. FEBS Lett. 579, 5385–5391 (2005).
Bilodeau, P. S. et al. The GAT domains of clathrin-associated GGA proteins have two ubiquitin binding motifs. J. Biol. Chem. 279, 54808–54816 (2004).
Kawasaki, M. et al. Molecular mechanism of ubiquitin recognition by GGA3 GAT domain. Genes Cells 10, 639–654 (2005).
Bennett, E. J. & Harper, J. W. DNA damage: ubiquitin marks the spot. Nature Struct. Mol. Biol. 15, 20–22 (2008).
Friedberg, E. C. et al. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol. Cell 18, 499–505 (2005).
Plosky, B. S. et al. Controlling the subcellular localization of DNA polymerases ι and ɛ via interactions with ubiquitin. EMBO J. 25, 2847–2855 (2006).
Guo, C. et al. Ubiquitin-binding motifs in REV1 protein are required for its role in the tolerance of DNA damage. Mol. Cell. Biol. 26, 8892–8900 (2006).
Crosetto, N. et al. Human Wrnip1 is localized in replication factories in a ubiquitin-binding zinc finger-dependent manner. J. Biol. Chem. 283, 35173–35185 (2008).
Meierhofer, D. et al. Quantitative analysis of global ubiquitination in HeLa cells by mass spectrometry. J. Proteome Res. 7, 4566–4576 (2008).
Xu, P. et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137, 133–145 (2009).
Hofmann, R. M. & Pickart, C. M. In vitro assembly and recognition of Lys-63 polyubiquitin chains. J. Biol. Chem. 276, 27936–27943 (2001).
Saeki, Y. et al. Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 28, 359–371 (2009).
Deveraux, Q. et al. A 26 S protease subunit that binds ubiquitin conjugates. J. Biol. Chem. 269, 7059–7061 (1994).
Lam, Y. A. et al. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature 416, 763–767 (2002).
Archer, C. T. et al. Physical and functional interactions of monoubiquitylated transactivators with the proteasome. J. Biol. Chem. 283, 21789–21798 (2008).
Hamazaki, J. et al. Rpn10-mediated degradation of ubiquitinated proteins is essential for mouse development. Mol. Cell. Biol. 27, 6629–6638 (2007).
Ng, J. M. et al. Developmental defects and male sterility in mice lacking the ubiquitin-like DNA repair gene mHR23B. Mol. Cell. Biol. 22, 1233–1245 (2002).
Young, P. et al. Characterization of two polyubiquitin binding sites in the 26S protease subunit 5a. J. Biol. Chem. 273, 5461–5467 (1998).
Zhang, N. et al. Structure of the S5a:K48-linked diubiquitin complex and its interactions with Rpn13. Mol. Cell 35, 280–290 (2009).
Jin, L. et al. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 133, 653–665 (2008).
Yao, T. et al. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nature Cell Biol. 8, 994–1002 (2006).
Qiu, X. B. et al. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 25, 5742–5753 (2006).
Hamazaki, J. et al. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 25, 4524–4536 (2006). References 80–82 demonstrate that RPN13 is a proteasome subunit that recruits the DUB UCH37 to the proteasome.
Lam, Y. A. et al. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature 385, 737–740 (1997).
Chen, L. & Madura, K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol. Cell. Biol. 22, 4902–4913 (2002).
Elsasser, S. et al. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J. Biol. Chem. 279, 26817–26822 (2004).
Saeki, Y. et al. Ubiquitin-like proteins and Rpn10 play cooperative roles in ubiquitin-dependent proteolysis. Biochem. Biophys. Res. Commun. 293, 986–992 (2002).
Kaplun, L. et al. The DNA damage-inducible UbL-UbA protein Ddi1 participates in Mec1-mediated degradation of Ho endonuclease. Mol. Cell. Biol. 25, 5355–5362 (2005).
Verma, R. et al. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell 118, 99–110 (2004).
Elsasser, S. et al. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nature Cell Biol. 4, 725–730 (2002).
Hiyama, H. et al. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26S proteasome. J. Biol. Chem. 274, 28019–28025 (1999).
Walters, K. J. et al. Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry 41, 1767–1777 (2002).
Matiuhin, Y. et al. Extraproteasomal Rpn10 restricts access of the polyubiquitin-binding protein Dsk2 to proteasome. Mol. Cell 32, 415–425 (2008). Extraproteasomal Rpn10 is shown to prevent toxicity caused by overexpression of Dsk2 by restricting its access to the proteasome.
Bertolaet, B. L. et al. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nature Struct. Biol. 8, 417–422 (2001).
Wang, Q. et al. Ubiquitin recognition by the DNA repair protein hHR23a. Biochemistry 42, 13529–13535 (2003).
Ortolan, T. G. et al. The DNA repair protein Rad23 is a negative regulator of multi-ubiquitin chain assembly. Nature Cell Biol. 2, 601–608 (2000).
Raasi, S. & Pickart, C. M. Rad23 UBA domains inhibit 26S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J. Biol. Chem. 278, 8951–8959 (2003).
Kang, Y. et al. Defining how ubiquitin receptors hHR23a and S5a bind polyubiquitin. J. Mol. Biol. 369, 168–176 (2007).
Fallon, L. et al. A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signalling. Nature Cell Biol. 8, 834–842 (2006).
Hoeller, D. et al. Regulation of ubiquitin-binding proteins by monoubiquitination. Nature Cell Biol. 8, 163–169 (2006).
Polo, S. et al. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416, 451–455 (2002).
Shih, S. C. et al. A ubiquitin-binding motif required for intramolecular monoubiquitylation, the CUE domain. EMBO J. 22, 1273–1281 (2003).
Hoeller, D. et al. E3-independent monoubiquitination of ubiquitin-binding proteins. Mol. Cell 26, 891–898 (2007). Shows that UBDs can utilize ubiquitin coupled to E2 enzymes to mediate the monoubiquitylation of host proteins independently of E3 ligase activity.
Woelk, T. et al. Molecular mechanisms of coupled monoubiquitination. Nature Cell Biol. 8, 1246–1254 (2006).
Crosetto, N. et al. Ubiquitin hubs in oncogenic networks. Mol. Cancer Res. 4, 899–904 (2006).
Hoeller, D. & Dikic, I. Targeting the ubiquitin system in cancer therapy. Nature 458, 438–444 (2009).
Verma, R. et al. Ubistatins inhibit proteasome-dependent degradation by binding the ubiquitin chain. Science 306, 117–120 (2004).
Cornilescu, G. et al. Validation of protein structure from anisotropic carbonyl chemical shifts in a dilute liquid crystalline phase. J. Am. Chem. Soc. 120, 6836–6837 (1998).
Zhang, D. et al. Affinity makes the difference: nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J. Mol. Biol. 377, 162–180 (2008).
Sato, Y. et al. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J. 28, 2461–2468 (2009). Structural determination of the Lys63 chain in complex with tandem UIMs, indicating that sequences between UIMs contribute to the specificity in Lys63–ubiquitin binding.
Fisher, R. D. et al. Structure and ubiquitin binding of the ubiquitin-interacting motif. J. Biol. Chem. 278, 28976–28984 (2003).
Hofmann, K. & Bucher, P. The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem. Sci. 21, 172–173 (1996).
Kirkin, V. et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33, 505–516 (2009).
Prag, G. et al. Mechanism of ubiquitin recognition by the CUE domain of Vps9p. Cell 113, 609–620 (2003).
Mizuno, E. et al. STAM proteins bind ubiquitinated proteins on the early endosome via the VHS domain and ubiquitin-interacting motif. Mol. Biol. Cell 14, 3675–3689 (2003).
Iha, H. et al. Inflammatory cardiac valvulitis in TAX1BP1-deficient mice through selective NF-κB activation. EMBO J. 27, 629–641 (2008).
Kanayama, A. et al. TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol. Cell 15, 535–548 (2004).
Meyer, H. H. et al. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1–Npl4. EMBO J. 21, 5645–5652 (2002).
Boyault, C. et al. HDAC6-p97/VCP controlled polyubiquitin chain turnover. EMBO J. 25, 3357–3366 (2006).
VanDemark, A. P. et al. Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell 105, 711–720 (2001).
Sundquist, W. I. et al. Ubiquitin recognition by the human TSG101 protein. Mol. Cell 13, 783–789 (2004).
Stamenova, S. D. et al. Ubiquitin binds to and regulates a subset of SH3 domains. Mol. Cell 25, 273–284 (2007). The first report that SH3 domains can non-covalently bind to ubiquitin.
Fu, Q. S. et al. Structural basis for ubiquitin recognition by a novel domain from human phospholipase A2 activating protein. J. Biol. Chem. 284, 19043–19052 (2009).
Bellare, P. et al. A role for ubiquitin in the spliceosome assembly pathway. Nature Struct. Mol. Biol. 15, 444–451 (2008).
Acknowledgements
We thank M. Bienko, N. Crosetto, D. Hoeller and D. McEwan for constructive comments and critical reading of the manuscript and the members of our laboratories for discussions. We thank X. Chen, N. Crosetto, M. Kawasaki, T. Kensche, S. Rahighi, P. Zhou and S. Skånland for help in making the figures. Research in the I.D. laboratory is supported by the Deutsche Forschungsgemeinschaft and the Cluster of Excellence “Macromolecular Complexes” of the Goethe University Frankfurt (EXC115), in the S.W. laboratory by the Target Protein Research Project of the MEXT, Japan, and in the K.J.W. laboratory by the US National Institutes of Health (CA097004 and CA117888) and the American Cancer Society (RSG-07-186-01-GMC).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Related links
Related links
DATABASES
FURTHER INFORMATION
Glossary
- 26S proteasome
-
A large protein complex that carries out regulated degradation to control protein lifespan. Proteasome activity is essential for a large range of cellular events, including cell cycle progression, DNA repair, apoptosis and the removal of misfolded proteins.
- Endosome
-
A small vesicle that is formed by endocytosis (invagination) of the plasma membrane, along with surface receptors, and is responsible for sorting internalized proteins and other biomolecules. Depending on the cargoes, endosomes are transported back to the cell surface for recycling, transported to the Golgi apparatus or matured into lysosomes.
- β-Sheet
-
A secondary structural element of proteins, in which the peptide backbone is almost fully extended.
- 310 Helix
-
A secondary structural element of proteins, in which a coiled conformation enables the formation of hydrogen bonds between backbone carbonyl and amide groups of amino acids that are three residues apart.
- α-Helix
-
A secondary structural element of proteins, in which a coiled conformation enables the formation of hydrogen bonds between backbone carbonyl and amide groups of amino acids that are four residues apart.
- Residual dipolar coupling
-
(RDC). Dipole–dipole coupling between spin 1/2 nuclei in samples that are partially aligned with an external magnetic field. RDC provides the orientation of bonds between neighbouring atoms relative to the rest of the molecule and can therefore be used in structure determination by NMR spectroscopy.
- Zinc finger
-
A small structural motif in proteins that is stabilized by interactions between amino acid side-chain atoms and a coordinated zinc atom.
- Ubiquitin conjugating (UBC) domain
-
The α/β structural fold of E2 conjugating enzymes that is characterized by a conserved active-site Cys residue, which forms a thiolester bond with ubiquitin.
- Plekstrin homology (PH) fold
-
A structural fold in proteins that is characterized by a distinct pattern of β-strands and an α-helix. This protein family tends to have a core of bulky hydrophobic amino acids.
- Ubiquitin-binding zinc finger
-
(UBZ). A subclass of ubiquitin-binding zinc finger domain that can bind to ubiquitin and control DNA damage responses.
- Ubiquitin-associated (UBA) domain
-
A small structural domain in proteins that is characterized by a three-helix bundle and is typically associated with the ubiquitin pathway.
- CUE domain
-
(Coupling of ubiquitin conjugation to endoplasmic reticulum degradation domain). A protein structural domain that is similar to the UBA domain. CUE domains form a three-helix bundle.
- ERAD
-
(Endoplasmic reticulum-associated degradation). A pathway by which misfolded proteins are transported from the endoplasmic reticulum to the 26S proteasome in the cytosol.
- Translesion synthesis polymerase
-
A member of a group of DNA polymerases that carry out translesion synthesis past DNA lesions. These polymerases, mostly belonging to the Y-family, have an open configuration and can accommodate different damaged bases in their active sites.
- UBAN domain
-
(Ubiquitin binding in ABIN and NEMO domain). An α-helical domain that is present in NEMO, ABINs and optineurin. It binds specifically to linear ubiquitin chains.
- Coiled coil
-
A structural motif in which α-helices coil around each other to enable favourable interactions between amino acid side-chain atoms.
- Multivesicular body
-
(MVB). A specialized endosome that is formed from early and sorting endosomes through invagination of the membrane enriched with surface receptors, resulting in many internalized vesicles. MVBs mature into late endosomes and lysosomes for degradation of internalized surface receptors.
- Ubiquitin-binding motif
-
(UBM). A structural ubiquitin-binding motif that is present in translesion DNA polymerases and is required for proper localization of these enzymes in nuclear replication foci.
- Peptidomimetic inhibitor
-
A chemical or natural compound that mimics a peptide–peptide interaction through a non-peptide bond or structure.
Rights and permissions
About this article
Cite this article
Dikic, I., Wakatsuki, S. & Walters, K. Ubiquitin-binding domains — from structures to functions. Nat Rev Mol Cell Biol 10, 659–671 (2009). https://doi.org/10.1038/nrm2767
Issue Date:
DOI: https://doi.org/10.1038/nrm2767
This article is cited by
-
Efficient determination of the accessible conformation space of multi-domain complexes based on EPR PELDOR data
Journal of Biomolecular NMR (2023)
-
Cav2.2-NFAT2-USP43 axis promotes invadopodia formation and breast cancer metastasis through cortactin stabilization
Cell Death & Disease (2022)
-
Structure of UBE2K–Ub/E3/polyUb reveals mechanisms of K48-linked Ub chain extension
Nature Chemical Biology (2022)
-
ANKRD13a controls early cell-death checkpoint by interacting with RIP1 independent of NF-κB
Cell Death & Differentiation (2022)
-
A context-dependent and disordered ubiquitin-binding motif
Cellular and Molecular Life Sciences (2022)