Abstract
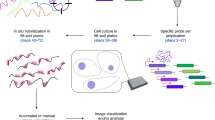
We describe a high-throughput protocol for RNA in situ hybridization (ISH) to Drosophila embryos in a 96-well format. cDNA or genomic DNA templates are amplified by PCR and then digoxigenin-labeled ribonucleotides are incorporated into antisense RNA probes by in vitro transcription. The quality of each probe is evaluated before ISH using a RNA probe quantification (dot blot) assay. RNA probes are hybridized to fixed, mixed-staged Drosophila embryos in 96-well plates. The resulting stained embryos can be examined and photographed immediately or stored at 4 °C for later analysis. Starting with fixed, staged embryos, the protocol takes 6 d from probe template production through hybridization. Preparation of fixed embryos requires a minimum of 2 weeks to collect embryos representing all stages. The method has been used to determine the expression patterns of over 6,000 genes throughout embryogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Gall, J.G. & Pardue, M.L. Formation and detection of RNA–DNA hybrid molecules in cytological preparations. Proc. Natl Acad. Sci. USA 63, 378–783 (1969).
Jones, K.W. & Robertson, F.W. Localisation of reiterated nucleotide sequences in Drosophila and mouse by in situ hybridisation of complementary RNA. Chromosoma 31, 331–345 (1970).
Harrison, P.R., Conkie, D., Paul, J. & Jones, K. Localisation of cellular globin messenger RNA by in situ hybridisation to complementary DNA. FEBS Lett. 32, 109–112 (1973).
Hafen, E., Levine, M., Garber, R. & Gehring, W. An improved in situ hybridization method for the detection of cellular RNAs in Drosophila tissue sections and its application for localizing transcripts of the homeotic Antennapedia gene complex. EMBO J. 2, 617–623 (1983).
Akam, M.E. The location of Ultrabithorax transcripts in Drosophila tissue sections. EMBO J. 2, 2075–2084 (1983).
Cowman, A.F., Zuker, C.S. & Rubin, G.M. An opsin gene expressed in only one photoreceptor cell type of the Drosophila eye. Cell 44, 705–710 (1986).
Banerjee, U., Renfranz, P.J., Pollock, J.A. & Benzer, S. Molecular characterization and expression of sevenless, a gene involved in neuronal pattern formation in the Drosophila eye. Cell 49, 281–291 (1987).
Tautz, D. & Pfeifle, C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98, 81–85 (1989).
Lehmann, R. & Tautz, D. In situ hybridization to RNA. Methods Cell Biol. 44, 575–598 (1994).
Kopczynski, C.C. et al. A high throughput screen to identify secreted and transmembrane proteins involved in Drosophila embryogenesis. Proc. Natl Acad. Sci. USA 95, 9973–9978 (1998).
Simin, K. et al. Profiling patterned transcripts in Drosophila embryos. Genome Res. 12, 1040–1047 (2002).
Tomancak, P. et al. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 3, RESEARCH0088 (2002).
Tomancak, P. et al. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 8, R145 (2007).
Rubin, G.M. et al. A Drosophila complementary DNA resource. Science 287, 2222–2224 (2000).
Stapleton, M. et al. The Drosophila gene collection: identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res. 12, 1294–1300 (2002).
Stapleton, M. et al. A Drosophila full-length cDNA resource. Genome Biol. 3, research0080 (2002).
Eugene, O., Yund, A. & Fristrom, J.W. Preparative isolation and short term organ culture of imaginal discs of Drosophila melanogaster . Tissue Culture Assoc. Manual 5, 1055–1062 (1979).
Kornberg, T., Siden, I., O'Farrell, P. & Simon, M. The engrailed locus of Drosophila: in situ localization of transcripts reveals compartment-specific expression. Cell 40, 45–53 (1985).
Schroder, C., Tautz, D., Seifert, E. & Jackle, H. Differential regulation of the two transcripts from the Drosophila gap segmentation gene hunchback. EMBO J. 7, 2881–2887 (1988).
Jazwinska, A., Kirov, N., Wieschaus, E., Roth, S. & Rushlow, C. The Drosophila gene brinker reveals a novel mechanism of Dpp target gene regulation. Cell 96, 563–573 (1999).
Pfeiffer, B.D. et al. Tools for neuroanatomy and neurogenetics in Drosophila . Proc. Natl. Acad. Sci. USA 105, 9715–9720 (2008).
Harmon, C.L. et al. Comparative analysis of spatial patterns of gene expression in Drosophila melanogaster imaginal discs. In Research in Computational Molecular Biology: Proc. 11th Annual Intl. Conf. (RECOMB 2007), Lecture Notes in Bioinformatics 4453 (Eds. Speed, T. and Huang, H.), 533–547 (Springer-Verlag, Berlin, Germany, 2007).
Lecuyer, E. et al. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131, 174–187 (2007).
Fowlkes, C.C. et al. A quantitative spatiotemporal atlas of gene expression in the Drosophila blastoderm. Cell 133, 364–774 (2008).
Acknowledgements
We thank current and former members of the BDGP, especially Amy Beaton, Joe Carlson, Erwin Frise, Elaine Kwan, Todd Laverty, Barret Pfeiffer, Stephen Richards, Mark Stapleton, Pavel Tomancak and Gerry Rubin for their contributions to these protocols as part of the systematic determination of patterns of gene expression in the Drosophila embryogenesis project. We thank Ken Wan for critical comments on the manuscript. This work was supported by the NIH Grant R01 GM076655 (SEC) through the US Department of Energy under contract no. DE-AC02-05CH11231.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Weiszmann, R., Hammonds, A. & Celniker, S. Determination of gene expression patterns using high-throughput RNA in situ hybridization to whole-mount Drosophila embryos. Nat Protoc 4, 605–618 (2009). https://doi.org/10.1038/nprot.2009.55
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2009.55
This article is cited by
-
An open-source semi-automated robotics pipeline for embryo immunohistochemistry
Scientific Reports (2021)
-
Synthesis and biological roles of O-glycans in insects
Glycoconjugate Journal (2020)
-
Optimized RNA ISH, RNA FISH and protein-RNA double labeling (IF/FISH) in Drosophila ovaries
Nature Protocols (2013)
-
Dual fluorescence detection of protein and RNA in Drosophila tissues
Nature Protocols (2012)
-
Systematic image‐driven analysis of the spatialDrosophilaembryonic expression landscape
Molecular Systems Biology (2010)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.