Abstract
Mutations in mismatch repair genes lead to Lynch Syndrome, the most common form of inherited colorectal cancer. In this report, we describe a novel complex germline mutation c.[1601_1661+92dup; 1591_1611del] of the mismatch repair gene, MSH2. This mutation, which segregates with the disease phenotype, was discovered in a Lynch syndrome kindred that also shows a history of the Muir-Torre syndrome. Interestingly, several tumors from this family displayed microsatellite instability, a hallmark of Lynch syndrome tumors but no consistent, concomitant loss of MSH2 protein expression. In addition, a subset of tumors showed neither prototypical feature of microsatellite instability nor immunohistochemistry deficiency, highlighting the importance of a detailed molecular analysis of rare genetic alterations. This mutation and the atypical clinical manifestations observed underscore the genetic complexity underlying Lynch syndrome, and the importance of comprehensive molecular screening in the diagnosis and early detection of colorectal and other associated cancers.
Similar content being viewed by others
Introduction
Lynch syndrome or hereditary nonpolyposis colorectal cancer (HNPCC; MIM 120435), is the most common colorectal cancer predisposition syndrome. It is an autosomal dominant disorder characterized by the development of cancers in the colorectum and endometrium, and to a lesser extent, cancers of the stomach, urinary tract, ovaries, small bowel and brain. Colorectal tumors from Lynch syndrome patients tend to be located in the proximal colon, have a better prognosis than sporadic colorectal tumors and are associated with distinct histological characteristics such as tumor-infiltrating lymphocytes, mucinous/signet-ring differentiation and/or a medullary growth pattern.
Germline mutations of the mismatch repair (MMR) genes MLH1 (MIM 120436), MSH2 (MIM 609309), PMS2 (MIM 600259) and MSH6 (MIM 600678) have hitherto been implicated in the Lynch syndrome. The MMR pathway has a central role in maintaining genomic stability by repairing DNA replication errors and signaling DNA damage caused by a variety of agents. Defective MMR results in microsatellite instability (MSI), characterized by an expansion or contraction of the number of tandem repeats that occur across the genome. MSI is a hallmark feature of Lynch syndrome, and is seen in about 85% of these tumors.1
Germline MMR defects also lead to Muir-Torre syndrome (MIM no. 158320). Muir-Torre is characterized by tumors in the sebaceous glands or keratoacanthomas that are associated with one or more of the various visceral neoplasms, mainly colorectal, endometrial, urological and upper gastrointestinal neoplasms. Muir-Torre syndrome is associated with germline mutations in the MSH2 gene, and to a lesser extent with mutations in MLH1 and MSH6 genes.2, 3, 4, 5
The human MSH2 gene is composed of 16 exons that span 934 amino acids. Similar to the other MMR genes, diverse sequence variants ranging from large insertions and deletions to missense mutations have been reported throughout the coding region of the MSH2 gene, and many of these are cataloged in the LOVD and MMR Variant Databases (http://chromium.liacs.nl/LOVD2/colon_cancer/home.php and http://www.med.mun.ca/MMRvariants).6
In this report, we describe a novel complex MSH2 germline mutation c.[1601_1661+92dup; 1591_1611del] identified in a family, which shows both Lynch and Muir-Torre syndromes. The unique nature of this mutation, together with several interesting clinical features seen in this family prompted us to carry out a detailed in vitro and in silico analysis of this mutation. Our analysis underscores the genetic complexity underlying cancer predisposition syndromes such as Lynch syndrome, and the importance of comprehensive molecular screening of patients/families eligible for diagnostic testing procedures.
Materials and methods
Selection of patients and families
Members of this kindred were referred to the molecular genetics laboratories as part of the Provincial Cancer Genetics Program for the assessment of the possible diagnosis of Lynch syndrome. Predictive genetic testing was offered to clinically affected and at risk subjects, with pre- and post-test genetic counseling as described previously.7 In addition to complete follow-up information, clinical and histopathological data were collected retrospectively on all affected patients. Informed consent was obtained from all subjects and all studies were performed according to guidelines of the Ethics Committee of the University of Toronto.
Microsatellite instability and immunohistochemistry
Microsatellite instability (MSI) testing and immunohistochemical (IHC) analysis for MMR proteins were performed as described previously.8 Matched normal and tumor DNA were assessed using a panel of up to five microsatellite markers as recommended by the National Cancer Institute.9 Each case was designated as either microsatellite unstable (MSI-H; ⩾30% markers unstable), microsatellite low (MSI-L; <30% markers unstable), or microsatellite stable (MSS; no unstable markers). IHC analysis of the respective MMR proteins was performed on formalin-fixed, paraffin-embedded tissues.
Mutation detection
Lymphocytes were isolated from blood samples using NH4Cl–Tris. DNA was extracted from lymphocytes using the Qiagen or saturated salt-out method as described previously.10 DNA from patients, whose tumors showed deficiency for an MMR protein, was subjected to exon by exon sequencing of genomic DNA to screen for alterations in MLH1 and MSH2 on an ABI 377 DNA Sequencer (Applied Biosystems, Foster City, CA, USA). Exons 7 and 13 of MYH were also screened for the two common mutations Y179C and G396D (formerly Y165C and G382D). Sequence information of the coding region was derived from RefSeq NM_000249.2 (MLH1), NM_000251.1 (MSH2) and NM_001128425.1 (MYH). Sequence information regarding the intronic regions was derived from GenBank U41215.1 (exon 10) and U41216.1 (exon 11). PCR conditions and primer sequences are available on request.
Reverse transcription PCR
RNA extraction was performed using TRIzol according to the manufacturer's protocol (Invitrogen, Burlington, ON, USA). Reverse transcription PCR was performed according to standard techniques as described before.10 To determine the sequence of RT-PCR products, the band of interest was excised and DNA was extracted by use of the QIAquick Gel Extraction Kit followed by gel electrophoresis (Qiagen, Mississauga, ON, USA). The DNA extracted was then sequenced according to the manufacturer's protocol.
Computational methods
The RepeatMasker (http://www.repeatmasker.org/) software was used to determine if sequences homologous to short or long interspersed elements (SINEs/LINEs) were present in the regions of the complex rearrangement. To assess for tandem and inverted repeats the programs Mreps (http://mobyle.pasteur.fr/cgi-bin/MobylePortal/portal.py?form=mreps) and Palindrome (http://mobyle.pasteur.fr/cgi-bin/MobylePortal/portal.py?form=palindrome) were used. Default settings were used for all programs.
Results
Clinical presentation and family history
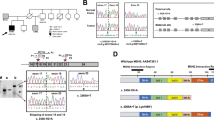
The proband identified in our study was diagnosed with cancer of the recto-sigmoid colon at the age of 50. His colon tumor was found to be MSI-H, but showed intact expression of all four MMR proteins by IHC. This patient was later, at the age of 61, diagnosed with a sebaceous adenoma on his forehead and a clear-cell renal carcinoma of the right kidney. The clear-cell tumor was found to be MSS, as well as intact by IHC for all of the MMR proteins. The proband's family history met Amsterdam I criteria (Figure 1a). The tumor characteristics and age of diagnosis for all known family members are summarized in Table 1 The co-occurrence of sebaceous gland cancers, as well as keratoacanthomas with internal neoplasms typical of the Lynch spectrum is consistent with a diagnosis of Muir-Torre syndrome in this family.
(a) Pedigree of the family of Proband 1: Solid symbols indicate individuals affected with cancer and open symbols indicate unaffected individuals. The proband is indicated by a solid arrow. dx: age at diagnosis; d: age at death. (b) Illustration depicting the complex mutation in exon 10 of MSH2.The horizontal line indicates the normal DNA sequence. The dashed ellipse above indicates the deleted (del) region and the solid box in line with the coding sequence indicates the duplicated (dup) region. The numbers depict the nucleotide sequence of the regions that are deleted/inserted. The figure is not drawn to scale.
In vitro molecular characterization
Based on these observations this patient's DNA was screened for underlying mutations in the MSH2 gene. Exon by exon sequencing combined with long range PCR analysis revealed a 153 bp duplication of the region from c.1601 and continuing until 1661+92. Following this duplication, a 21 bp fragment from c.1591–1611 is deleted in MSH2 exon 10. The duplication event most likely happened before the deletion, because the duplication includes part of the deleted region (c.1601–1611). Based on this description this mutation can be referred to as c.[1601_1661+92dup; 1591_1611del] based on the most likely sequence of events that may have occurred. This complex mutation is illustrated in Figure 1b. RT-PCR analysis showed that the deletion of c.1591–1611 led to an in-frame 21 bp (seven amino acid) deletion in exon 10 of MSH2, and confirmed that the insertion in intron 10 was not part of the MSH2 transcript. Subsequent analysis has revealed that the proband's father, sister and daughter are also carriers of this complex germline mutation. The father's and sister's tumors are part of a spectrum of tumors seen in Lynch syndrome, confirming that the mutation segregates with the disease phenotype. Sequencing of the two MYH gene mutation hotspot regions also showed that the proband, his sister and his father were negative for the common mutations, Y179C and G396D (formerly described as Y165C and G382D).
Bioinformatic analysis
The region surrounding this mutation was analyzed for repetitive elements using the software RepeatMasker. Although no repetitive elements were found at the locations where the deletion or the duplication occurred, our analysis indicates that this mutation occurs downstream of a region that shows homology to the long interspersed nuclear element-1 (LINE-1/L1), L1P4 (c.1661+100 to 1661+194). To gain further insight into the molecular basis of this mutation, we carried out computer-based sequence analysis using Mreps and Palindrome to assess the region for tandem and inverted repeats respectively. No tandem repeats were identified in the vicinity of the mutation; however, the 5′ break point for the deletion occurred within a 10-bp inverted repeat sequence (c.1586–1595) that is palindromic to an upstream sequence in intron 9 (from c.1510–18 to 1510–27).
Discussion
We identified a complex mutation, c.[1601_1661+92dup; 1591_1611del] in exon 10 of MSH2 in this proband, that occurs upstream of an L1 sequence, LIP4. In addition, our analysis indicated that the 5′ break point occurs within a palindromic sequence.
LINEs have been implicated in several germline mutations (reviewed in Kazazian and Moran11). Previous reports document two large germline MLH1 deletions in which the break points mapped to an L1 element and nonrepetitive DNA sequence, respectively.12 Viel et al., reported a 2454-bp deletion in MLH1, likely because of the recombination between two L1 elements in intron 2 and intron 3.13 Interestingly, this allele with the MLH1 deletion was also characterized by a complex mutation delCCinsACATAGTA, which gave rise to a palindromic sequence in the proximity of the fusion site. It was proposed that the complex mutation delCCinsACATAGTA arose in the context of a repair process involving filing in and nick ligation as previously reported for the COL4A5 and COL4A6 genes in Alport syndrome.14 In fact, deletions in the COL4A5 and COL4A6 genes were shown to arise by a diverse mechanism from resident L1 repeats.14 Thus, it is plausible that the complex mutation in our proband arose as a result of a repair process that attempted to correct a large deletion deep within the intronic region. In addition, a previous report of a MLH1 complex mutation invoked a mechanism involving non-homologous end joining (NHEJ) during the repair of double-strand breaks.15 NHEJ is one of primary mechanisms by which eukaryotes repair double-strand breaks. However, given that NHEJ uses little or no sequence similarity to re-join ends, it is a process that is prone to many errors.16 Although there is no definitive evidence, it is also possible that this mutation arose in an attempt to repair a double-strand break using NHEJ. However, further studies are needed to precisely identify if NHEJ is involved in this process.
The discovery of a palindromic sequence at the deletion break point is interesting, as a previous analysis showed that palindromic sequences flank 5% of all the deletions and insertions studied in the p53 gene.17 Of the sequences inserted or deleted between palindromes, 40% were found to be greater than 20 bp, compared with 12% of mutations flanked by tandem repeats. Thus, while the exact mechanism that led to the formation of c.[1601_1661+92dup; 1591_1611del] is unclear, it is evident that the features of this stretch of DNA likely contributed to the formation of this complex mutation.
Several tumors of this kindred showed both MSI and MSS phenotypes. Furthermore, a subset showed MSI, but did not show MSH2 deficiency by IHC. This could be because the region recognized by the MSH2 antibody (c-terminal portion of the protein) was not sufficiently altered in these patients, consequently leading to an inconsistent immunohistochemical staining pattern. In addition, it is also possible that the in-frame deletion had minimal impact on the expression/stability of the protein. This is supported by the observation that this mutation maps on to the clamp domain of MSH2 based on the recently solved crystal structures of human MutSα (MSH2–MSH6 complex).18 Based on these structures, this clamp domain is postulated to be involved in making thus far uncharacterized, non-specific interactions with DNA. Therefore, this mutation likely decreases the efficiency with which MSH2 recognizes and binds to certain types of DNA mismatches, or the stability of these interactions; thereby affecting the efficient functioning of the MMR system, rather than the expression of the MSH2 protein. Another interesting feature of this family was the presence of tumors that were both MSS and intact for MMR gene expression by IHC. One possibility is that this complex MSH2 mutation did not affect MSH2 protein function and expression in a manner necessary to lead to MSI and/or loss of protein expression. Alternatively, it is possible that these tumors are sporadic in nature and not associated with the Muir-Torre/Lynch syndrome spectrum.
It has been approximated that 35% of tumors in individuals positive for the Muir-Torre Syndrome do not display instability of microsatellite repeats, which combined with other features of Muir-Torre has spurred the speculation that at least two distinct forms of Muir-Torre exist.19 The first subtype, which includes MSI-positive tumors, shares genetic and pathological features with Lynch syndrome. It is characterized by early-onset colorectal carcinoma and a strong family history of cancer. The second group of MSS or MMR-proficient tumors shows later ages of onset and a less pronounced family history, and is likely because of genes that are not implicated in MMR pathway. The base excision repair gene MYH has been proposed to be a likely candidate gene that is mutated in these families.19, 20 Biallelic inactivation of MYH can lead to an autosomal recessive form of inherited colorectal cancer known as MYH associate polyposis. A recent report describes the presence of sebaceous gland tumors in several individuals positive for germline MYH mutations.20 Our analysis indicated that neither the proband nor his clinically affected family members carried the common germline MYH mutations. Furthermore, given the segregation of a complex germline MSH2 mutation in this family and the presence of a subset of MSI-H tumors, it is not likely that their colorectal tumors belong to the MMR-proficient Muir-Torre category.
Another notable feature of this proband's family history is the presence of prostate adenocarcinoma. Although tumors of the urinary tract, specifically cancers of the renal pelvis and ureter are associated with HNPCC, prostate cancer is a relatively recent addition to the Lynch syndrome tumor spectrum. Prostate cancer has occasionally been described in HNPCC families and interestingly, the first observation linking MSI to prostate cancer was made in a Muir-Torre patient.21 A recent study published by the German HNPCC consortium, found a correlation between prostate cancers and MSH2 mutation carriers, leading them to propose that this should be taken into consideration during clinical and genetic counseling of MSH2 mutation carriers.22
In summary, the systematic characterization of this complex mutation in MSH2 using in vitro methods, followed by bioinformatic analysis has shed light on the complex molecular mechanisms underlying MMR gene mutations. The analysis of tumors from this kindred also highlights the challenges of using either IHC or MSI analysis alone, as an indicator of an underlying MMR gene defect. This is especially relevant as the diagnosis of Lynch syndrome relies primarily on the detection of germline defects in the MMR genes of these patients.23 In addition, this case illustrates the importance of carefully evaluating and documenting rare genetic variations and their associated clinical symptoms to better understand Muir-Torre syndrome and its link with Lynch syndrome. As underscored by several previous reports, a single sebaceous adenoma can be the only clue that tips off the presence of colon cancer and the inherited predisposition to cancer.24 Therefore, the systematic evaluation of families displaying Muir-Torre characteristics can have an important role in both the diagnosis and early detection of colon and other associated cancers.
References
Lynch, H. T. & de la Chapelle, A. Genetic susceptibility to non-polyposis colorectal cancer. J. Med. Genet. 36, 801–818 (1999).
Bapat, B., Xia, L., Madlensky, L., Mitri, A., Tonin, P., Narod, S. A. et al. The genetic basis of Muir-Torre syndrome includes the hMLH1 locus. Am. J. Hum. Genet. 59, 736–739 (1996).
Kruse, R., Lamberti, C., Wang, Y., Ruelfs, C., Bruns, A., Esche, C. et al. Is the mismatch repair deficient type of Muir-Torre syndrome confined to mutations in the hMSH2 gene? Hum. Genet. 98, 747–750 (1996).
Mangold, E., Rahner, N., Friedrichs, N., Buettner, R., Pagenstecher, C., Aretz, S. et al. MSH6 mutation in Muir-Torre syndrome: could this be a rare finding? Br. J. Dermatol. 156, 158–162 (2007).
Arnold, A., Payne, S., Fisher, S., Fricker, D., Soloway, J., White, S. M. et al. An individual with Muir-Torre syndrome found to have a pathogenic MSH6 gene mutation. Fam. Cancer. 6, 317–321 (2007).
Woods, M. O., Williams, P., Careen, A., Edwards, L., Bartlett, S., McLaughlin, J. R. et al. A new variant database for mismatch repair genes associated with Lynch syndrome. Hum. Mutat. 28, 669–673 (2007).
Soravia, C., Sugg, S. L., Berk, T., Mitri, A., Cheng, H., Gallinger, S. et al. Familial adenomatous polyposis-associated thyroid cancer: a clinical, pathological, and molecular genetics study. Am. J. Pathol. 154, 127–135 (1999).
Woods, M. O., Hyde, A. J., Curtis, F. K., Stuckless, S., Green, J. S., Pollett, A. F. et al. High frequency of hereditary colorectal cancer in Newfoundland likely involves novel susceptibility genes. Clin. Cancer. Res. 11, 6853–6861 (2005).
Rodriguez-Bigas, M. A., Boland, C. R., Hamilton, S. R., Henson, D. E., Jass, J. R., Khan, P. M. et al. A National Cancer Institute Workshop on Hereditary Nonpolyposis Colorectal Cancer Syndrome: meeting highlights and Bethesda guidelines. J. Natl. Cancer. Inst. 89, 1758–1762 (1997).
Charames, G. S., Ramyar, L., Mitri, A., Berk, T., Cheng, H., Jung, J. et al. A large novel deletion in the APC promoter region causes gene silencing and leads to classical familial adenomatous polyposis in a Manitoba Mennonite kindred. Hum. Genet. 124, 535–541 (2008).
Kazazian, Jr. H. H. & Moran, J. V. The impact of L1 retrotransposons on the human genome. Nat. Genet. 19, 19–24 (1998).
Li, L., McVety, S., Younan, R., Liang, P., Du Sart, D., Gordon, P. H. et al. Distinct patterns of germ-line deletions in MLH1 and MSH2: the implication of Alu repetitive element in the genetic etiology of Lynch syndrome (HNPCC). Hum. Mutat. 27, 388 (2006).
Viel, A., Petronzelli, F., Della Puppa, L., Lucci-Cordisco, E., Fornasarig, M., Pucciarelli, S. et al. Different molecular mechanisms underlie genomic deletions in the MLH1 Gene. Hum. Mutat. 20, 368–374 (2002).
Segal, Y., Peissel, B., Renieri, A., de Marchi, M., Ballabio, A., Pei, Y. et al. LINE-1 elements at the sites of molecular rearrangements in Alport syndrome-diffuse leiomyomatosis. Am. J. Hum. Genet. 64, 62–69 (1999).
McVety, S., Younan, R., Li, L., Gordon, P. H., Wong, N., Foulkes, W. D. et al. Novel genomic insertion—deletion in MLH1: possible mechanistic role for non-homologous end-joining DNA repair. Clin. Genet. 68, 234–238 (2005).
Kolomietz, E., Meyn, M. S., Pandita, A. & Squire, J. A. The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumors. Genes Chromosomes Cancer. 35, 97–112 (2002).
Greenblatt, M. S., Grollman, A. P. & Harris, C. C. Deletions and insertions in the p53 tumor suppressor gene in human cancers: confirmation of the DNA polymerase slippage/misalignment model. Cancer Res. 56, 2130–2136 (1996).
Warren, J. J., Pohlhaus, T. J., Changela, A., Iyer, R. R., Modrich, P. L. & Beese, L. S. Structure of the human MutSalpha DNA lesion recognition complex. Mol Cell. 26, 579–592 (2007).
Ponti, G. & Ponz de Leon, M. Muir-Torre syndrome. Lancet Oncol. 6, 980–987 (2005).
Ponti, G., Ponz de Leon, M., Maffei, S., Pedroni, M., Losi, L., Di Gregorio, C. et al. Attenuated familial adenomatous polyposis and Muir-Torre syndrome linked to qcompound biallelic constitutional MYH gene mutations. Clin. Genet. 68, 442–447 (2005).
Honchel, R., Halling, K. C., Schaid, D. J., Pittelkow, M. & Thibodeau, S. N. Microsatellite instability in Muir-Torre syndrome. Cancer Res. 54, 1159–1163 (1994).
Goecke, T., Schulmann, K., Engel, C., Holinski-Feder, E., Pagenstecher, C., Schackert, H. K. et al. Genotype-phenotype comparison of German MLH1 and MSH2 mutation carriers clinically affected with Lynch syndrome: a report by the German HNPCC Consortium. J Clin. Oncol. 24, 4285–4292 (2006).
Casey, G., Lindor, N. M., Papadopoulos, N., Thibodeau, S. N., Moskow, J., Steelman, S. et al. Conversion analysis for mutation detection in MLH1 and MSH2 in patients with colorectal cancer. JAMA 293, 799–809 (2005).
Rothenberg, J., Lambert, W. C., Vail, Jr. J. T., Nemlick, A. S. & Schwartz, R. A. The Muir-Torre (Torre's) syndrome: the significance of a solitary sebaceous tumor. J. Am. Acad. Dermatol. 23, 638–640 (1990).
Acknowledgements
We thank the patients and their family members for their participation in this study. We also gratefully acknowledge the assistance provided by the staff of the Molecular Diagnostics Clinic, Mt Sinai Hospital, and the Credit Valley Hospital. We thank George Charames for helpful discussions. SP is a recipient of the Canadian Institutes for Health Research (CIHR)/Canadian Institute for Digestive Health Doctoral Award and the Frank Fletcher Memorial award from the University of Toronto.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Perera, S., Ramyar, L., Mitri, A. et al. A novel complex mutation in MSH2 contributes to both Muir-Torre and Lynch Syndrome. J Hum Genet 55, 37–41 (2010). https://doi.org/10.1038/jhg.2009.119
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2009.119