Abstract
Like many insects, mosquitoes, rely on endosymbionts to grow and develop. These can be acquired from the environment. We used next generation 454 pyrosequencing to discern the whole-body microbiome of the mosquito species Culex quinquefasciatus in various larval stadia and following exposure to common pharmaceutical and personal care products (PPCPs) found in wastewater. PPCP treatments included environmentally-relevant concentrations; 1) a combination of common antibiotics, 2) a combination of mammalian hormones, 3) a mixture of the antibiotic and hormone treatments plus acetaminophen and caffeine and, 4) an untreated control. Within control groups, the predominant families of bacterial symbionts change with each larval instar despite consistent diets and rearing conditions. This trend was also seen in hormone treatments but not in the antibiotic or the mixture treatments. Richness and evenness were reduced in both antibiotic and mixture treatments, suggesting that antibiotics remove certain bacteria or inhibit them from increasing to proportions seen in the control treatment. Interestingly, the mixture treatments had greater richness and evenness compared to antibiotic alone treatments, possibly due to the other contaminants facilitating growth of different bacteria. These findings illuminate the complexity of the microbiome of C. quinquefasciatus and may have implications for more effective control strategies.
Similar content being viewed by others
Introduction
Endosymbionts, bacterial species known to grow, develop, and vertically transmit in an organisms’ cells, usually for mutualistic symbiosis, are essential to the growth, development, and fecundity of many insect species1,2,3,4,5. Many aphid species, such as Acyrthosiphon pisum, Megoura viciae, and Myzus persicae, rely heavily on endosymbionts to survive on the unbalanced diet of phloem sap. Their bacteriocyte endosymbiont, Buchnera spp, provides essential amino acids6. Without Buchnera, aphids demonstrate reduced growth rates and development and produce few or no offspring6,7. However, some endosymbionts in the genus Wolbachia are known to manipulate insects for their own benefit and can also lead to increased vector competency for transmission of human diseases such as West Nile Virus8,9. Wolbachia species are common vertically transmitted endosymbionts in many mosquito species and typically infect reproductive tissue10. There are many species of Wolbachia that influence a variety of insect behaviours and life-history traits. Some endosymbionts can also act defensively by killing parasitoid eggs after they are laid in the host8,10,11. Because endosymbionts play major roles in insect development, they have been proposed for use in insect control12.
Due to the importance of endosymbionts for development in many insect species, some species vertically transfer endosymbionts (from parent to offspring). For example, Estes et al.13 describes the microbiome of dung beetles’ (Onthophagus taurus) brood balls, which are used to nourish their offspring until they are adults. When the microbiota in the beetle offspring and their female parent were compared in sterilized dung and soil, they had proportionally identical 16S rRNA sequences from their microbiome. Interestingly, over their life stages, the proportions of the families of bacteria in the beetles’ microbiome changed. The predominant family of the first three instars varied by individual. However, from the pupal stage onwards, Enterobacteriaceae was the most common family in the dung beetle. Bees acquire important bacteria through social interaction and also from the environment14. Without some of these bacteria, it is thought that honey bees could become more susceptible to outside diseases and increase incidents of colony collapse15. More studies are needed to fully understand the importance of microbiota in mosquitoes as they have been linked to increased transmission of pathogens from mosquitoes to humans9,16.
Mosquitoes are common disease vectors, which spend their juvenile stages in aquatic environments17. Bacteria from the water, both symbiotic and free-living, have been shown to influence the microbiome of mosquitoes, suggesting that some of their possible endosymbionts are collected from the environment18. Consequently, if the environment is altered, and some of these necessary bacteria are reduced/eliminated, there could be detrimental effects on the development of mosquito larvae. Such a removal effect may occur as the result of antibiotic runoff or other pollution, and/or environmental changes. For example, Rosi-Marshall et al.19 showed that common pharmaceuticals in streams will alter the respiration and diversity of biofilms. Pennington et al.20 reported differences in whole-body microbiomes and increased developmental times for Culex quinquefasiatus (Say) treated with various pharmaceuticals and personal care products (PPCPs).
Chemicals intended for human use often occur in aquatic environments and/or enter water supplies through water treatment plant overflow or from use of reclaimed water in water scarce areas21,22,23,24,25,26. These chemicals can then affect bacterial communities in the water and the associated aquatic insect community. Presence of these contaminants can alter effectiveness of Bacillus thuringiensis subsp. israelensis (Bti) a pesticide commonly applied for mosquito control20. However, little is known about how such contaminants will influence the microbiome of such insects. Similarly, there is a lack of data determining if mosquito bacterial communities vary during the course of larval development; all available studies we are aware of examine only late instar larvae or mixed lower instars and species5,27, and information regarding mosquitoes’ endosymbionts and their function is very limited or non-existent outside of Wolbachia. Therefore, we describe the differences in the bacterial communities of the mosquito C. quinquefasciatus in multiple distinct larval stages, as well as when these mosquitoes are reared in environments contaminated with ecologically relevant concentrations of PPCPs that commonly occur in combinations in order to provide a baseline for more in-depth studies.
Results
The mosquitoes’ microbiomes were significantly affected by both PPCPs (PERMANOVA: F = 3.78; df = 3, 32; p < 0.001) and instar (PERMANOVA: F = 8.64; df = 2, 33; p < 0.001). Additionally, there was a significant interaction of PPCP treatment and instars (PERMANOVA: F = 7.63; df = 6, 29; p < 0.001). The microbiome of the control mosquitoes changed significantly between instars (PERMANOVA: F = 10.39; df = 2, 8; p < 0.01). In pairwise comparisons of only larvae from the control treatments, second instar larvae were significantly different from third instar (p < 0.01) but were not different than fourth instars (p = 0.1) while, third and fourth instars were not significantly different (p = 0.1014) from each other. Families with at least 1% proportionality of the control treated mosquitoes’ microbiome in at least one instar were also examined via pairwise comparisons. The results are included in Table 1.
When PPCP treatments were examined for differences among instars, second (PERMANOVA: F = 4.05; df = 3, 8; p < 0.001), third (PERMANOVA: F = 10.25; df = 3, 8; p < 0.001), and fourth (PERMANOVA: F = 9.63; df = 3, 8; p < 0.001), were significantly affected by PPCP treatments. When subjected to pairwise comparisons, antibiotic and hormone treated mosquitoes’ microbiomes were significantly different between second and fourth instar only (p < 0.01), and there was no significant differences in any instars in the mixture treated mosquitoes (p = 0.3736).
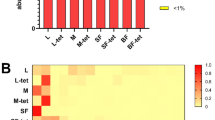
PCA (Fig. 1) demonstrates that various instars separate from each other in the four treatment groups with third and fourth instar loading similarly on the first dimension and all three instars loading distinctly in the second dimension. Bacterial families Oxalobacteraceae and Aeromonadaceae closely follow the separation pattern in the first dimension and Cryomorphaceae follows in the second (Table 2). In the antibiotic treatments second instar loaded separately from third and fourth on the second dimension (Fig. 1). The bacterial family Propionibacteriaceae also follows this trend and was the only family with at least 85% correlation in either of the first two dimensions (Table 2). For hormone treated mosquitoes, second instars loaded separately from fourth instars on the first dimension (Fig. 1). The bacterial families, which follow this trend, are Enterobacteriaceae and Pseudomonadaceae. As shown in Fig. 2 and Table 2, control treatments’ bacterial families changed with instar; starting with Cytophagaceae (56.25%) in second instars, changing to Enterobacteriaceae (41.81%) in third instar and finally the control fourth instars’ most predominant family was Microbacteriaceae (34.89%). There were a total of eight bacterial families with proportionalities greater then 1% in at least one instar of the control treatment. In fourth instars there is a resurgence of bacterial families from second instars, which were overshadowed by the third instar bacterial families (Table 1). Interestingly, the family Rickettsiaceae was the second most predominant family in all control instars. Operational taxonomic units (OTUs) assigned to the family Rickettsiaceae were found in most treatments and instars, although they were reduced in many hormone treated samples relative to the controls and increased in the antibiotic treatments. Notably, antibiotic and mixture treatment groups’ most predominant family was Rickettsiaceae over all instars. The second most predominant family in antibiotic and mixture treatment groups was Sphingobacteriaceae in all instars. Hormone treatments changed bacterial communities between instars but not as drastically as the control group. Second instars exposed to hormones predominately contained Oxalobacteraceae, which changed to Microbacteriaceae and Rickettsiaceae in the third instar. The predominant family of fourth instar hormones was Microbacteriaceae, although some proportion of Rickettsiaceae was still present.
In the alpha diversity analysis, richness (number of different species in one location) was examined as mean observed species (Fig. 3) and evenness (distribution of species’ proportionalities in one location) was measured by mean Shannon’s index (Fig. 4). For mean observed species at a sequencing depth of 3000 sequences/sample there was a significant difference between treatments (Χ2: 870.40; df: 3; p < 0.001), instars (Χ2: 80.02; df: 2; p < 0.001) and a significant interaction of treatment and instar (Χ2: 136.76; df: 6; p < 0.001). Mosquitoes treated with antibiotics had lower richness and fewer total sequences per sample than all other treatments with the richness decreasing as larvae age. This is evident in Fig. 2, as there are proportionally fewer bacterial families outside of Rickettsiaceae and Sphingobacteriaceae than in other treatments. In contrast, mosquitoes reared in the mixture of hormones, antibiotics and the common contaminants, acetaminophen and caffeine; demonstrate a relatively constant richness over time (Fig. 3). The control groups and hormone treatments fluctuate more than the mixture and antibiotic treatment groups but demonstrate consistently higher richness (Fig. 4). The mean Shannon’s diversity index suggests that antibiotics alone substantially reduced diversity. The mixture treatments also display reduced diversity, however, they are more diverse than their antibiotic treatment counterparts. The control groups display a greater diversity than both the antibiotic alone and mixture treatments when compared by increasing instar, where as the hormone treatment group, had no discernable pattern. The mixture also displays no discernable pattern compared by instar, which is likely due to the effects of the hormones added to antibiotics. Finally, it is notable that in some treatments, the mean species number (Fig. 3) failed to reach an asymptote. This may indicate that an inadequate sample size for detecting extremely rare species, or that the reduced diversity of some samples allowed for the entire community to be described.
Discussion
Here we have demonstrated that the microbiome of larval Culex mosquitoes changes throughout development, and variation between instars is affected when exposed to various PPCPs. It has previously been demonstrated that mosquitoes rely on their microbiomes to aid in development and that removing certain symbionts can significantly slow larval development5,28,29. Pennington et al.20 demonstrated that PPCPs at environmentally relevant concentrations, which are significantly lower than those used in most laboratory studies, can alter the microbiome of mosquitoes and slow their development.
In the field, Duguma et al.27 showed pooled Culex species’ microbiomes will change from early (first and second) to late (third and fourth) instars. Coon et al.5 and Wang et al.29 showed that the microbiome of mosquitoes will change as the insects advance from fourth instar larvae, to pupae, to the adult stage, and after adults fed on a blood meal. We have shown that the microbiome of early (second and third) instars’ will change from one instar to the next even without exposure to PPCPs (control group). In the second, third, and fourth instars, predominant families change from Cytophagaceae to Enterobacteriaceae and finally to Microbacteriaceae. However similar to Pennington et al.20, third and fourth instar were not significantly different and our findings also correlate to what Duguma et al.27 found in their laboratory reared Culex tarsalis late instars. However, as their third and fourth instars were pooled, only the Enterobacteriaceae family predominates. These families were all removed in the antibiotic and mixture treatments. Fourth instar larvae in the control group match what was described in Aedes aegypti by Coon et al.5. Coon et al.5 also described the microbiome of two other mosquito species (Anopheles gambiae and Georgecraigius atropalpus) during the fourth instar. Their microbiomes had different proportions of familial microbiota between each other and both were different from the findings in our C. quinquefasciatus fourth instars. To our knowledge ours is the first study to look at the microbiome changes of individual early instars in mosquitoes. This suggests the possibility of a new strategy for mosquito control targeting the critical microorganisms essential for development at specific stages. Specifically, additional research targeting key symbionts found in earlier instars would determine if the younger larvae can be controlled more effectively, as has been seen with pesticides such as Bti30.
A number of mosquitoes are common carriers of the bacterial genus Wolbachia, which usually acts as a reproductive parasite in the ovaries of the females, and is suspected to be in at least 20% of all insect species31. As in Pennington et al.20, Rickettsiaceae, the family containing Wolbachia pipientis, continuously holds the majority count of the antibiotic and mixture treatments’ microbiome. When the OTUs mapped to the family Rickettsiaceae was examined at the level of genus the predominant and sole genus detected was Wolbachia. Rickettsiaceae is vertically transmitted from mother to offspring32; however, for many of the other bacterial families present, it is difficult to discern the source or how they are incorporated into the insects’ microbiome. This is made further complicated since these traits may vary by species or genus and mapping OTUs to finer taxonomic levels was generally not possible. Similarly, it may be possible to determine some origins via comparison with the water in rearing pans over time, although there was no DNA found in water at the start of the experiments. However, our focus was not on the origin of bacterial species in these mosquitoes, and we do not have these data. Analyses or the microbial community in such pans would be an interesting follow-up study. Interestingly, Enterobacteriaceae, which includes the genus Buchnera and other common endosymbionts, is the predominant family of the third instars in the control treatment. For example, the gut symbiont of the plataspid stinkbug (Megacopta punctatiss) is phylogenetically similar to Buchnera species33. In potato psyllids (Bactericera cockerelli) various genera of the family Enterobacteriaceae have been reported in the life stages and faeces accounting for at least 21% of the microbiome34,35. Enterobacteriaceae is one of, if not the most important family of endosymbionts in the pea aphid (Acyrthosiphon pisum)6, and is commonly used in research regarding the effects of antibiotics on insect-symbiont interactions7. Similarly, Chouaia et al. (2012)28 described a slowing of larval development in Anopheles mosquitoes when they removed Asaia bacteria from the family Acetobacteraceae. However this family has one of the lowest proportionalities in all of the mosquitoes including control treated. This suggests it is not an endosymbiont of this Culex mosquito species. However, reports of the effects of other PPCPs on insect-symbiont interactions are rare.
It is interesting that the hormones found in wastewater from treatment plants (and in the hormone treatment groups) are all mammalian female sex hormones and would not be expected to affect bacteria. We would not expect an effect of these hormones on bacteria, as they have no endocrine system; nonetheless, substantial changes in the microbiome occurred in response to exposure to these hormones (Fig. 1). Similarly, caffeine and an antihistamine, would not be expected to effect biofilms, but were shown to repress respiration in stream biofilms19. We think there may be some influence the hormones have on bacterial gene expression however that is not in the scope of this paper and thus, specific effects for each PPCP or combination of contaminants will need to be determined from more experimental data.
The increased bacterial diversity during mosquito ontogeny could result from either bacterial replication during development or by acquisition through ingestion. In the hormone and control treatment groups, we are unsure if bacteria are lost during development, or if the change in bacterial diversity was caused by differential growth among taxa. Interestingly, in pairwise comparisons of the hormone treated mosquitoes (Table 2), the majority of the significant differences were between second and fourth instar larvae. The hormone treated mosquitoes also had the most families that were correlated to the first principal component at a minimum of 85%. Combined this suggests that the mosquitoes exposed to hormones had the most diverse microbial communities and that this diversity increases over time.
Mammalian hormones change the microbiome of C. quinquefasciatus mosquitoes and it is possible they are responsible for the increased richness and diversity seen in the mixture treatments compared to antibiotics alone; however more studies will need to be conducted to confirm this conclusion. Regardless, our results indicate that reclaimed wastewater has the potential to impact mosquito ecology. Considerably more research will be required to discern how mixtures of PPCPs could affect bacterial microbiomes for important medical pests. If similar results are found for agriculturally important insects (including either pests and/or beneficial insects) exposed to these emerging contaminants, additional research documenting the effects of increasing use of reclaimed water and associated changes to the insect microbiome will become even more important. Similarly, because insects are a critical food source for higher trophic level organisms in terrestrial surface waters, releases of PPCPs in aquatic environments have the potential to modify the ecology of these ecosystems.
Materials and Methods
Insect Rearing
Culex quinquefasciatus mosquito egg rafts from colonies continuously reared at the University of California, Riverside, CA were maintained using the methods of Wirth et al.36. Egg rafts were held in shallow porcelain pans (30 × 20 × 5 cm) containing 3 L Geyser® Natural Alpine Spring Water (C G Roxane, Olancha, CA) or spring water and one of three PPCP treatments (described below). All pans contained food and any biologic/inorganic materials from the food and mosquito waste. It was beyond the scope of this project to simulate all of the possible small concentrations of various organic materials that could appear in wastewater from different treatment plants. Thus, the growing environments were made as reproducible as possible while still containing organics from the diet. Egg rafts and larvae were maintained in an incubator (model 818: Precision Scientific Inc., Buffalo, NY) at 28 °C, approximately 70% RH, and a light: dark cycle of 16:8. Mosquitoes were fed 2 mL of a mixture of 4 g brewers yeast and ground mouse chow (1:3 wt: wt) rehydrated in 50 mL water every 3 d. PPCP concentrations (Table 3) were used as in Pennington et al.20, and based on concentrations found by Kolpin et al.21 and Mutiyar and Mittal37. The treatment groups examined were: a control consisting of only water, an antibiotic treatment of lincomycin, oxytetracycline, and ciprofloxacin (Alfa Aesar, Ward Hill, MA; purity ≥98%), a hormone treatment of estrone, 19-norethindrone, 17β- estradiol, and 17α- ethynylestradiol (Sigma-Aldrich, St. Louis, MO; purity ≥98%), and a mixture of all PPCPs plus acetaminophen (MP Biomedicals, LLC, Santa Ana, CA; purity ≥90%) and caffeine (Fisher Scientific, Hanover Park, IL; laboratory grade purity). Despite Pennington et al.20 demonstrating that acetaminophen and caffeine do not alter the microbiome, these were included in the mixture treatment because they can and do co-occur in wastewater with the other contaminants and we did not want to assume there were no possible joint effects. The extended data set showing acetaminophen and caffeine has been presented as Supplementary Fig. S1. Hydrochloric acid (Fisher Scientific; 12.1 M) and sodium hydroxide pellets (Sigma-Aldrich, St. Louis, MO) were used to prepare 1 M stock solutions for adjusting final pH in rearing pans to 7 ± 0.5.
Extraction of Bacterial DNA
In preparation for sequencing, three mosquitoes from each PPCP treatment were collected as second, third and fourth instars, as first instars were too small to collect without damage, and twice washed with 95% ethanol to remove any external microorganisms. Larvae were then transferred to individual sterile 2 mL microcentrifuge tubes with 95% ethanol and stored at −60 ± 3 °C in an ultra cold freezer (Forma Scientific, Inc. Marietta, OH) until DNA extraction. DNA was extracted using a Qiagen DNeasy® Blood and Tissue Kit following the manufacturers protocols amended as in Pennington et al.20. In addition to mosquitoes samples of water and water plus diet were also extracted using identical protocols, with the additional step of centrifugation at 2900 rpm in an IEC HN-SII tabletop centrifuge for 1 h to create a pellet. Upon extraction, nucleic acid concentration was quantified using a Nanodrop ND- 2000c Spectrophotometer (Cole-Palmer, Vernon Hills, IL), to confirm enough genetic material for sequencing. This process revealed no DNA in water or water and diet samples when no mosquitoes were present and thus they were not subjected to further analysis because any bacteria would have originated from the mosquitoes, their egg-rafts, or from the air after the water had been altered by the mosquitoes various biological processes.
Roche 454 bacteria barcoded amplicon pyrosequencing was performed by a commercial sequencing facility (Molecular Research LP MR DNA, Shallowater, TX). The procedure used the forward primer 27Fmod (GRGTTTGATCMTGGCTCAG) and the reverse primer 519Rmodbio (GTNTTACNGCGGCKGCTG) in a single-step 30 cycle PCR using HotStarTaq Plus Master Mix (Qiagen, Valencia, CA). PCR was performed using the following cycle conditions: 94 °C for 3 min, followed by 28 cycles of 94 °C for 30 s; 53 °C for 40 s, 72 °C for 1 min; and a final elongation at 72 °C for 5 min. After PCR, all amplicons were mixed in equal concentrations and purified using Agencourt Ampure beads (Agencourt Bioscience Corporation, MA). Samples were then sequenced with Roche 454 FLX titanium instruments and reagents following the manufacturer’s guidelines.
Sequence Analysis Pipeline
Raw bacterial DNA sequences were analysed using a MacQIIME (version: 1.8.0-20140103) based pipeline38. Raw .fasta and .qual files were combined to .fastaq file by the convert_fastaqual_fastaq.py command and uploaded to NIH/NLM/NCBI Sequence Read Archive (SRA) under SRP study accession number: SRP067136. Barcodes and primers were trimmed using the split_libraries.py script with default settings. After barcode trimming, data was denoised and re-inflated using the default denoise_wrapper.py script settings and the default setting in the inflate_denoiser_output.py script. Following denoising, no samples contained any ambiguous nucleotides. The maximum, minimum, and mean number of sequences across all samples was 22128, 3102, and 10099.11 respectively, with the average length of the reads being 403.84 base pairs. Operational taxonomic units (OTUs) were chosen by the default 97% identity threshold, which roughly correlates to species39, via the UCLUST method as implemented in the pick_otus.py script40. Representative OTUs were chosen using the pick_rep_set.py script and default settings. The Greengenes reference database clustered at 97% identity was used to assign taxonomy using the assign_taxonomy.py script41. OTUs were counted and summarized using the make_otu_table.py and summarize_taxa.py scripts respectively. OTUs were aligned using the align_seqs.py and filter_alignment.py scripts, and used to build a phylogenetic tree (make_phylogeny.py). There were 658 distinct OTUs at the species level with 58 distinct families; 15 OTUs failed to match any contained within the database and could not be assigned taxonomically. Fifteen families were chosen by their proportionality being greater than or equal to 1% in at least one sample for the heatmap. The cut-off was chosen at 1% as this was assumed to be the minimum to influence larval development at that stage. For alpha diversity, multiple rarefactions were performed using the multiple_rarefaction.py script with the lowest rarefaction depth of 2000, the highest rarefaction depth of 21000, a step size of 1000, and a replicate number of ten, which normalizes the data at each depth. Alpha diversity was calculated using the alpha_diversity.py script with the metrics observed species (species richness) and Shannon Indices (evenness) from the raw data40,42,43. Alpha diversity data was not averaged between replicate mosquitoes as they have been averaged by resampling-replicates and the complications and validity of this is still being considered44,45. Metrics were summarized using the collate_alpha.py script.
Statistical Analyses
Statistical analyses were performed using R (the R Foundation for Statistical Computing, version 3.1.1). Following processing through the QIIME pipeline, “Permutational MANOVA” (PERMANOVA) in the Vegan package46 was used to compare the OTU data (Supplementary dataset 1). Independent variables were instar (n = 3), PPCP treatment (n = 4) and the interaction of the two, with three replicates (n = 3) of each instar in each PPCP treatment and control (n = 36). PERMANOVA is analogous to MANOVA but is suited to address the non-normality that is commonly associated with count data in ecological community and genetic data47,48. Microbial community data were further examined via principal component analysis (PCA) performed in the FactoMineR package49. Ellipses in the PCA encompass the three mosquito replicates in each instar for that treatment. PCA and PERMANOVA were conducted on each instar in the individual PPCP and control treatment groups. Following PCA, variables were examined for their contributions and correlation to each of the first two dimensions. Those variables (OTUs) that were ≥85% correlated were included in subsequent pairwise comparisons by instar in their respective treatment. Generalized linear hypotheses was used to perform pairwise comparisons in the multcomp package50. P values were adjusted using the p.adjust command. Alpha diversity data was analysed using a negative binomial generalized linear models at a sequence depth of 3000 sequences/sample to normalize data to the highest number where all sample mosquitoes were present. The alpha level for all tests was 0.05.
Additional Information
How to cite this article: Pennington, M. J. et al. Culex quinquefasciatus larval microbiomes vary with instar and exposure to common wastewater contaminants. Sci. Rep. 6, 21969; doi: 10.1038/srep21969 (2016).
References
Kuechler, S. M., Gibbs, G., Burckhardt, D., Dettner, K. & Hartung, V. Diversity of bacterial endosymbionts and bacteria-host co-evolution in Gondwanan relict moss bugs (Hemiptera: Coleorrhyncha: Peloridiidae). Environ. Microbiol. 15, 2031–42 (2013).
Nachappa, P., Levy, J., Pierson, E. & Tamborindeguy, C. Diversity of endosymbionts in the potato psyllid, Bactericera cockerelli (Triozidae), vector of zebra chip disease of potato. Curr. Microbiol. 62, 1510–20 (2011).
Kafil, M., Bandani, A. R., Kaltenpoth, M., Goldansaz, S. H. & Alavi, S. M. Role of symbiotic bacteria in the growth and development of the Sunn pest, Eurygaster integriceps . J. Insect Sci. 13, 1–12 (2013).
Minard, G., Mavingui, P. & Moro, C. V. Diversity and function of bacterial microbiota in the mosquito holobiont. Parasit. Vectors 6, 1–12 (2013).
Coon, K. L., Vogel, K. J., Brown, M. R. & Strand, M. R. Mosquitoes rely on their gut microbiota for development. Mol. Ecol. 23, 2727–39 (2014).
Douglas, A. E. Nutritional interactions in insect-microbial symbioses: Aphids and their symbiotic bacteria Buchnera . Annu. Rev. Entomol. 43, 17–37 (1998).
Wilkinson, T. The elimination of intracellular microorganisms from insects: an analysis of antibiotic-treatment in the pea aphid (Acyrthosiphon pisum). Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 119, 871–881 (1998).
Clark, E. L., Karley, A. J. & Hubbard, S. F. Insect endosymbionts: manipulators of insect herbivore trophic interactions? Protoplasma 244, 25–51 (2010).
Dodson, B. L. et al. Wolbachia enhances West Nile Virus (WNV) infection in the mosquito Culex tarsalis . PLoS Negl. Trop. Dis. 8, 1–7 (2014).
Mouton, L. et al. Virulence, multiple infections and regulation of symbiotic population in the Wolbachia-Asobara tabida symbiosis. Genetics 168, 181–9 (2004).
Pannebakker, B. A., Loppin, B., Elemans, C. P. H., Humblot, L. & Vavre, F. Parasitic inhibition of cell death facilitates symbiosis. Proc. Natl. Acad. Sci. USA 104, 213–5 (2007).
Douglas, A. E. Symbiotic microorganisms: untapped resources for insect pest control. Trends Biotechnol. 25, 338–42 (2007).
Estes, A. M. et al. Brood ball-mediated transmission of microbiome members in the dung beetle, Onthophagus taurus (Coleoptera: Scarabaeidae). PLoS One 8, e79061 (2013).
McFrederick, Q. S. et al. Environment or kin: whence do bees obtain acidophilic bacteria? Mol. Ecol. 21, 1754–68 (2012).
Hamdi, C. et al. Gut microbiome dysbiosis and honeybee health. J. Appl. Entomol. 135, 524–33 (2011).
Hughes, G. L., Vega-Rodriguez, J., Xue, P. & Rasgon, J. L. Wolbachia strain wAlbB enhances infection by the rodent malaria parasite Plasmodium berghei in Anopheles gambiae mosquitoes. Appl. Environ. Microbiol. 78, 1491–95 (2012).
Becker, N. et al. Mosquiteos and their control. (Kluwer Academic/Plenum Publishers, 2003).
Demisse, D. Influence of Nutrients and Integrated Mosquito Management Tactics on Mosquitoes and Their Habitat Microbiomes. (ProQuest LLC, 2014).
Rosi-Marshall, E. J. et al. Pharmaceuticals suppress algal growth and microbial respiration and alter bacterial communities in stream biofilms. Ecol. Appl. 23, 583–93 (2013).
Pennington, M. J., Rivas, N. G., Prager, S. M., Walton, W. E. & Trumble, J. T. Pharmaceuticals and personal care products alter the holobiome and development of a medically important mosquito. Environ. Pollut. 203, 199–207 (2015).
Kolpin, D. W. et al. Pharmaceuticals, hormones, and other organic wastewater contaminants in US streams, 1999–2000: A national reconnaissance. Environ. Sci. Technol. 36, 1202–11 (2002).
Phillips, P. J. et al. Combined sewer overflows: an environmental source of hormones and wastewater micropollutants. Environ. Sci. Technol. 46, 5336–43 (2012).
Li, W. C. Occurrence, sources, and fate of pharmaceuticals in aquatic environment and soil. Environ. Pollut. 187, 193–201 (2014).
Carballa, M. et al. Behavior of pharmaceuticals, cosmetics and hormones in a sewage treatment plant. Water Res. 38, 2918–26 (2004).
Küster, A. et al. Environmental risk assessment of human pharmaceuticals in the European Union: A case study with the β-blocker Atenolol. Integr. Environ. Assess. Manag. 6, 514–23 (2009).
Chen, Y. et al. Occurrence and environmental implications of pharmaceuticals in Chinese municipal sewage sludge. Chemosphere 93, 1765–72 (2013).
Duguma, D., Hall, M. W., Rugman-Jones, P., Stouthamer, R. & Terenius, O. Developmental succession of the microbiome of Culex mosquitoes. BMC Microbiol. 1–13 (2015), 10.1186/s12866-015-0475-8.
Chouaia, B. et al. Delayed larval development in Anopheles mosquitoes deprived of Asaia bacterial symbionts. BMC Microbiol. 12 Suppl 1 (2012).
Wang, Y., Gilbreath, T. M., Kukutla, P., Yan, G. & Xu, J. Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PLoS One 6, e24767 (2011).
Van Essen, F. W. & Hembree, S. C. Laboratory bioassay of Bacillus thuringiensis israelensis against all instars of Aedes aegypti and Aedes taeniorhynchus larvae. Mosq. News 40, 424–31 (1980).
Werren, J. H. & Windsor, D. M. Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc. R. Soc. London 267, 1277–85 (2000).
McMeniman, C. J. et al. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti . Science. 323, 141–4 (2009).
Fukatsu, T. & Hosokawa, T. Capsule-transmitted gut symbiotic bacterium of the Japanese common plataspid stinkbug, Megacopta punctatissima . Appl. Environ. Microbiol. 68, 389–96 (2002).
Hail, D., Dowd, S. E. & Bextine, B. Identification and location of symbionts associated with potato psyllid (Bactericera cockerelli) lifestages. Environ. Entomol. 41, 98–107 (2012).
Arp, A., Munyaneza, J. E., Crosslin, J. M., Trumble, J. & Bextine, B. A global comparison of Bactericera cockerelli (Hemiptera: Triozidae) microbial communities. Environ. Entomol. 43, 344–52 (2014).
Wirth, M. C., Delécluse, A. & Walton, W. E. Laboratory selection for resistance to Bacillus thuringiensis subsp. jegathesan or a component toxin, Cry11B, in Culex quinquefasciatus (Diptera: Culicidae). J. Med. Entomol. 41, 435–41 (2004).
Mutiyar, P. K. & Mittal, A. K. Risk assessment of antibiotic residues in different water matrices in India: key issues and challenges. Environ. Sci. Pollut. Res. Int. 21, 7723–36 (2014).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Ong, S. H. et al. Species identification and profiling of complex microbial communities using shotgun illumina sequencing of 16S rRNA amplicon sequences. PLoS One 8, 1–8 (2013).
Kuczynski, J. et al. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Microbiol. 1E–5 (2012), 10.1002/9780471729259.mc01e05s27.
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–61 (2010).
Montagna, M. et al. Metamicrobiomics in herbivore beetles of the genus Cryptocephalus (Chrysomelidae): Towards the understanding of ecological determinants in insect symbiosis. Insect Sci. 340–52 (2014), 10.1111/1744-7917.12143.
Jing, X. et al. The bacterial communities in plant phloem-sap-feeding insects. Mol. Ecol. 23, 1433–44 (2014).
Richardson, J. M. L. & Richards, M. H. A randomisation program to compare species-richness values. Insect Conserv. Divers. 1, 135–141 (2008).
Crist, T. O., Veech, J. a., Gering, J. C. & Summerville, K. S. Partitioning species diversity across landscapes and regions: a hierarchical analysis of alpha, beta, and gamma diversity. Am. Nat. 162, 734–743 (2003).
Oksanen, J. et al. The Vegan Package. 1–190 (2008) at < http://cran.r-project.org/, http://vegan.r-forge.r-project.org/>.
Anderson, M. J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46 (2001).
McArdle, B. H. & Anderson, M. J. Fitting multivariate models to community data: A comment on distance-based redundancy analysis. Ecology 82, 290–7 (2001).
Husson, F., Josse, J., Le, S. & Mazet, J. FactoMineR: Multivariate exploratory data analysis and data mining with R. R Packag. version 1.14 102–23 (2010) at < http://www.cran.r-project.org/package=FactoMineR.fr>.
Hothorn, T. et al. Simultaneous Inference in General Parametric Models. Packag. ‘multcomp’ (2015) at < http://cran.stat.sfu.ca/web/packages/multcomp/multcomp.pdf>.
Acknowledgements
We would like to thank M. Wirth for supplying mosquito egg rafts and expertise on mosquito rearing. B. Vindiola, H. Drew, K. Gilbert, and N. Rivas aided in treating mosquitoes. Finally, we would like to thank, D. De La Riva, N. Di, J. Gan, K. Hladun, G. Kund, Q. McFrederick, R. Stouthamer, B. Vindiola, and K. Zheng, for providing comments on earlier versions of this manuscript.
Author information
Authors and Affiliations
Contributions
M.J.P., S.M.P. and J.T.T. designed the experiments and wrote the manuscript. M.J.P. and S.M.P. conducted the analyses. M.J.P. generated all figures and tables. The J.T.T. laboratory and the University of California, Riverside Environmental Toxicology Program funded the project. W.E.W. provided advice, insects, and rearing expertise. All authors reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Pennington, M., Prager, S., Walton, W. et al. Culex quinquefasciatus larval microbiomes vary with instar and exposure to common wastewater contaminants. Sci Rep 6, 21969 (2016). https://doi.org/10.1038/srep21969
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep21969
This article is cited by
-
Impact of Human Activities on Disease-Spreading Mosquitoes in Urban Areas
Journal of Urban Health (2023)
-
Estrogen induces shift in abundances of specific groups of the coral microbiome
Scientific Reports (2021)
-
Generational conservation of composition and diversity of field-acquired midgut microbiota in Anopheles gambiae (sensu lato) during colonization in the laboratory
Parasites & Vectors (2019)
-
The mosquito holobiont: fresh insight into mosquito-microbiota interactions
Microbiome (2018)
-
Changes in Larval Mosquito Microbiota Reveal Non-target Effects of Insecticide Treatments in Hurricane-Created Habitats
Microbial Ecology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.