Abstract
Manganese (Mn) is both an essential biological cofactor and neurotoxicant. Disruption of Mn biology in the basal ganglia has been implicated in the pathogenesis of neurodegenerative disorders, such as parkinsonism and Huntington's disease. Handling of other essential metals (e.g. iron and zinc) occurs via complex intracellular signaling networks that link metal detection and transport systems. However, beyond several non-selective transporters, little is known about the intracellular processes regulating neuronal Mn homeostasis. We hypothesized that small molecules that modulate intracellular Mn could provide insight into cell-level Mn regulatory mechanisms. We performed a high throughput screen of 40,167 small molecules for modifiers of cellular Mn content in a mouse striatal neuron cell line. Following stringent validation assays and chemical informatics, we obtained a chemical ‘toolbox’ of 41 small molecules with diverse structure-activity relationships that can alter intracellular Mn levels under biologically relevant Mn exposures. We utilized this toolbox to test for differential regulation of Mn handling in human floor-plate lineage dopaminergic neurons, a lineage especially vulnerable to environmental Mn exposure. We report differential Mn accumulation between developmental stages and stage-specific differences in the Mn-altering activity of individual small molecules. This work demonstrates cell-level regulation of Mn content across neuronal differentiation.
Similar content being viewed by others
Introduction
Most essential metals are highly regulated throughout development, serving as required co-factors while also implicated in cytotoxic processes1. Metal homeostasis is maintained through concerted mechanisms involving metal transporters, metallochaperone proteins, metal responsive signaling pathways and transcription factors and sequestration of metals within sub-cellular organelles2. A complex regulatory biology is known to underlie the transport and handling of most essential metals including iron, copper and zinc3,4,5. However, little is known about intracellular handling of Mn, despite it being an essential cofactor of cellular processes that rely on Mn-dependent enzymes such as superoxide dismutase (Mn-SOD), arginase, glutamine synthetase and neurotransmitter synthetic enzymes6. In excess, Mn is neurotoxic, producing a variety of motor and psychiatric disturbances that can be attributed to basal ganglia dysfunction7,8,9,10 and an enhanced susceptibility to Mn exposure has been reported during fetal and infant development11,12. Mn preferentially accumulates and elicits toxicity within basal ganglia structures such as the caudate, putamen, substantia nigra and globus pallidus13. Specifically, Mn has been implicated in parkinsonism and Huntington's disease, two neurodegenerative syndromes that show neuronal loss in the basal ganglia14,15,16,17,18.
Mn transport into the central nervous system has been well characterized at the level of the blood-brain barrier (BBB), consisting of facilitated and active transport through divalent metal transporter 1 (DMT1), transferrin receptor (TfR), ZIP819,20,21 and other transporters. However, the role of these and other metal transporters in neuronal Mn transport is not well understood22. Indeed, while a number of transporters (e.g. SLC30A10, ATP13A2, SPCA1 and SPCA2) contribute to Mn detoxification and other transporters have been identified as contributing to Mn uptake (e.g. DMT1, ZIP8 and ZIP14), none of these are selective to Mn and most have been studied only under extracellular Mn concentrations well above the levels of brain Mn (60 µM–150 µM) associated with neurotoxicity in vivo22,23. Mn-specific binding proteins or signaling pathways responsive to physiological Mn concentrations have not been described.
Because optimal intracellular Mn concentrations likely change over developmental time and lineage, we postulated that neurons and perhaps other cells, have cell-level homeostatic processes that regulate intracellular Mn content to cover cellular physiological demands while preventing cytotoxicity. Furthermore, utilization of distinct cellular Mn handling mechanisms may vary as physiological demand and availability of this co-factor change. Such regulatory processes may consist of similar mechanisms that have been identified for other essential metals3,4,5, including: intracellular Mn sensors, Mn chaperones or storage proteins and cell signaling systems with transcriptional and post-transcriptional control processes to regulate the balance of Mn across intracellular compartments. Identification of neuronal Mn handling processes has proven rather intractable by genetic and biochemical approaches, perhaps in part because phenotypes associated with insufficient or excess Mn are not clearly defined. We hypothesized that Mn handling mechanisms can be identified and later studied, by using a novel chemical biology approach that relies on intracellular Mn levels as the outcome measure. To identify small molecules that modify cellular Mn content, we performed a high throughput screen for modifiers of intracellular Mn status under biologically relevant levels of Mn. We then utilized these small molecules to test the hypothesis that cellular mechanisms of Mn handling are developmentally regulated between the highly relevant human floor-plate lineage neuroprogenitors and their derivative mesencephalic dopaminergic neurons10,24,25.
Results
High throughput screen identifies small molecules for manipulating neuronal Mn status
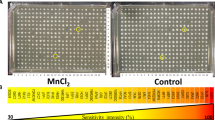
Our laboratory had previously developed the Cellular Fura-2 Mn Extraction Assay (CFMEA) to enable rapid fluorescent-based measurements of intracellular Mn content26. Here, we utilized CFMEA for high throughput screening (HTS). The overall HTS approach consisted of pretreatment with the test small molecule, followed by co-exposure to 125 µM MnCl2, representative of in vivo brain Mn levels at which aberrant function would be observed23. An immortalized murine striatal neuron lineage (STHdhQ7/Q7) was used in the HTS, as the striatum, like other structures of the basal ganglia, has a relatively high basal Mn level and an especially high capacity for Mn accumulation26,27. After the Mn and small molecule incubation period, extracellular Mn was washed away and CFMEA performed (Fig. 1a). During optimization of the assay for 384-well format, we determined the major sources of intra-plate variability to be volumetric delivery of fura-2 extraction buffer. We developed a strategy to control for variability in delivered fura-2 by supplementing the fura-2 extraction buffer with a dextran-coupled fluorophore (Alexa-568) that does not interfere with fluorometric readings of fura-2. This enabled us to correct for variations in robotic volumetric delivery (achieving Z’>0.5) (Supplementary Fig. S1). These and other optimizations enabled detection of changes in fura-2 signal as low as 10%, corresponding to a z-score of ± 3.0.
HTS identifies modulators of neuronal Mn status.
(a) HTS screening protocol. Timeline of screening paradigm indicating cell plating, 10 µM small molecule addition, Mn addition, washing, fura-2 extraction, signal measurement and analysis. (b) Composition of 40,167 screened small molecules. Individual small molecule libraries are labeled and total compounds in that collection are indicated. (c) HTS paradigm. Each stage of the screen and number of compounds refined at that stage. (d) Scatterplot of primary screen. Z-score of each small molecule is indicated (n = 1, per compound). A hit was defined as a small molecule with a z-score or b-score (not shown) cutoff of ± 3.0 or larger, as indicated by dotted line (green).
To comprehensively identify small molecules that modulate neuronal Mn levels, we screened a total of 40,167 small molecules spanning multiple chemical libraries (Fig. 1b). Of these, 901 (2.2% hit rate) small molecules increased or decreased Mn levels (z-score or b-score > ±3.0) (Fig. 1c,d). These hits were first validated by replication in duplicate followed by generation of 1 nM-10 µM concentration response curves (CRCs). A total of 133 small molecules (0.33%) passed these criteria, displaying a variety of potencies and patterns of activity (Supplementary Fig. S2). We then culled the toolbox of small molecules not readily obtainable from commercial or academic sources, leaving 65 for subsequent study. We further refined the toolbox by excluding fluorescent small molecules that could give false positive increases in fura-2 signal (Supplementary Fig. S3) and molecules that altered the number of cells under the conditions of the HTS screen due to cytotoxicity or other mechanisms (Supplementary Fig. S4). These validation and quality control measures resulted in a final Mn toolbox of 41 small molecules. The functional properties of this toolbox consisted of significantly more Mn level increasing small molecules than Mn level decreasing small molecules (Supplementary Table S1) (Chi-square, p<0.05). Given the tendency of small molecules from pharmacological screens to inhibit (antagonistic) rather than enhance (agonistic) the function of their biological target28, this skewing toward Mn level increasing small molecules suggests Mn efflux mechanisms are more susceptible to pharmacological disruption than mechanisms of Mn influx.
High structural diversity of validated small molecules
We generated a heat map of pairwise Tanimoto coefficients (Fig. 2), a measure of chemical structure similarity, using an upper triangle distance matrix considering largest common substructure between two molecules. As the majority of our identified small molecules have no known biological targets, we sought to reveal potential relationships between their chemical structure by performing hierarchical agglomerative clustering using complete linkage to generate functional dendrograms. This analysis revealed a high degree of structural diversity, with an average pairwise Tanimoto coefficient of 0.19, where a 1.0 translates to identical compounds and a 0.0 to compounds that have no common substructure. While three clusters of 2–4 similar molecules (Tanimoto coefficients >0.8) exist, most had little structural similarity with other active molecules (Tanimoto coefficient <0.5)29. This finding suggests that the screen did not identify the complete range of structure-activity relationships, likely reflecting both the diversity of biological targets that influence cellular Mn status, as well as the potential of diverse structures to influence common targets. The identified small molecule clusters all had Mn level increasing activity (Cluster 1 (VU0002852 and VU0003765), Cluster 2 (VU0088990, VU0029414), Cluster 3 (VU0027832, VU0028040, VU0028386, VU0133141)).
Structure activity dendrogram and Tanimoto plot of Mn toolbox.
Functional dendrogram with clustering based on chemical similarity as determined by Tanimoto score (1 = identical, 0 = unique). Small molecule activity pattern per node is indicated by color: Mn level decreasing small molecules (red), Mn level increasing small molecules (blue) and mixed activity (green). Compound Tanimoto scores for each small molecule were plotted across all compounds in the Mn toolbox represented by heat map.
Activity of small molecules at normal brain Mn levels
In the screen, small molecules were identified using a concentration of Mn associated with mammalian neurotoxicity in vivo (125 µM). Subsequently, we sought to assess the Mn modifying activity of these small molecules under normal brain Mn levels. Although in vivo concentrations of Mn may not directly translate to in vitro exposures, it has been shown that normal levels of human brain Mn range from 20 µM to 53 µM23. Thus, we tested for activity at 31.25 µM Mn, a four-fold dilution from the concentration used in the primary screen, that is also just above the minimum limit of Mn detection by CFMEA. The vast majority of small molecules (39/41, 95%) exhibited Mn-modifying activity at this Mn concentration (Fig. 3). There was considerable variation in effect size between our high and low Mn exposure conditions, with some small molecules having greater activity (fold change of Mn) at 125 µM Mn and others having greater activity at 31.25 µM Mn. Two molecules failed to have significant activity at 31.25 µM (VU0243195 and VU0482834). One molecule, VU0076546, had a unique pattern of activity, increasing intracellular neuronal Mn levels at the high Mn concentration, but strongly decreasing intracellular Mn levels at the lower Mn concentration.
Activity of validated small molecules at physiological and toxicological Mn exposures.
Fold change in Mn levels at both 31.25 µM (blue) and 125 µM (red) MnCl2 is plotted versus small molecule identity. Small molecules tested in hiPSC-derived mesencephalic floor plate neuroprogenitors and post-mitotic neurons are marked in (purple). ‘NS’ indicates small molecules that do not show significant activity for 31.25 µM concentration. Horizontal lines indicate the detection limit of CFMEA (green) and no effect on Mn levels (grey). Data plotted as mean ± SEM (n = 16 for 31.25 µM, n = 2 for 125 µM). Statistical significance (all not marked by NS) was determined by comparing small molecule versus vehicle by one-way ANOVA, p<0.05.
Chemical informatics reveals common pathways among predicted targets
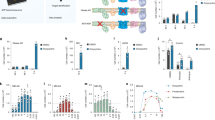
We performed a chemical informatics analysis of the 41 small molecule toolbox to identify potential biological targets. In order to reveal functional relationships between small molecules, we aggregated information regarding known targets of each molecule and its similar chemical homologs (Tanimoto coefficient >0.7). The structures of the entire Mn toolbox were inputted into STITCH 4.0 analysis generating two major signaling node clusters, with only 5 of the 41 chemicals having structural homologs with known or suspected biological activity. VU0482585, VU0239513, VU0482834 and VU0243195 exhibited multi-nodal interactions with a network comprised of mitogen-activated protein kinase 14 (MAPK14), transforming growth factor beta receptor I (TGFBR1), glutathione S-transferase P (GSTP1), superoxide dismutase 1 (SOD1), copper-transporting ATPase 1 (ATP7A), ATP-binding cassette sub-family C member 2 (ABCC2) and ATP-binding cassette sub-family G member 2 (ABCG2) (Fig. 4a). These predicted interactions were further supported by the fact that VU0482585 and VU0482834 were from the GSK Published Kinase Inhibitor Set30, suggesting a mechanism related to kinase inhibition or disruption of ATP-dependent transporters. In addition, homologs of VU0003610 have known binding to butyrylcholinesterase (BCHE) and acetylcholinesterase (ACHE) (Fig. 4b). While these findings provide some insight into potential biological targets, the majority of our identified small molecules and their close chemical homologs, do not have other published biological activities. This suggests that the neuronal Mn status modifying activity of our validated small molecules is relatively specific since such structures have not been identified in other published screens performed on the same publically available chemical libraries.
Chemical informatics analysis of Mn toolbox.
(a,b) STITCH analysis of Mn toolbox small molecules. Each node represents a small molecule or target protein. Color coded asterisk indicates corresponding small molecule or small molecule homolog (Tanimoto score > 0.7) validated in the HTS, identified in the legend. Color of connections between nodes indicates known function and description of the type of evidence available: activation (green), inhibition (red), binding (blue), catalysis (purple) and expression (yellow).
Differential regulation of Mn content in developing mesencephalic dopaminergic neurons
We hypothesized that small molecules in our Mn-modifying chemical toolbox would exhibit differential activity across neurodevelopmental time as the biological requirements for Mn change. We tested this hypothesis in developing human mesencephalic (floor plate lineage) dopaminergic neurons, given this lineage's particular susceptibility to Mn toxicity and involvement in neurodegenerative syndromes such as parkinsonism10,24,25. Human induced pluripotent stem cells (hiPSCs) generated from a control subject were differentiated into FOXA2+, LMX1A+ floor-plate lineage neuroprogenitors (12 days differentiation) and further to Tyrosine hydroxylase (TH)+, β3-Tubulin+, LMX1A+ post-mitotic early dopaminergic neurons (26 days differentiation) (Fig. 5a). Mn content was quantified by CFMEA following 125 µM MnCl2 exposure31,32. Early dopaminergic neurons (day 26) accumulated significantly (p<0.05) more intracellular Mn than the more immature floor plate neuroprogenitors (day 12) (Fig. 5b). Nine small molecules of varying structures and effect sizes were selected to evaluate their activity in this human system and test for potential developmental stage specificity. All tested small molecules (9 of 9) showed activity in hiPSC-derived mesencephalic dopaminergic neuroprogenitors (one-way ANOVA, p<0.001). Three of the small molecules (3 of 9, 33%) showed similar magnitude of activity at both neurodevelopmental time points (Fig. 6a–c). Four of the small molecules (4 of 9, 44%) exhibited greater activity in floor plate neuroprogenitors than post-mitotic dopaminergic neurons (Fig. 6d–g). Finally, two small molecules showed developmental-stage specific activity (2 of 9, 22%) (Fig. 6h,i). VU0047355, a negative modulator of Mn concentration and VU0482585, a positive modulator of Mn concentration, exhibited a significant impact on intracellular Mn levels exclusively in the more mature dopaminergic neurons, the developmental time point with the highest Mn accumulation between the two stages of differentiation (Fig. 6h,i). These results demonstrate that the validated small molecules act on developmentally regulated targets involved in control of intracellular neuronal Mn content.
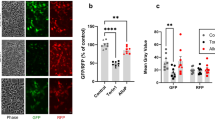
Mn accumulation changes across developmental time in mesencephalic floor-plate neuroprogenitors and/or post-mitotic dopaminergic neurons.
(a) Immunocytochemistry of day 12 floor plate neuroprogenitors and day 26 post-mitotic dopaminergic neurons. Appropriate developmental stage markers indicated by legend. (b) Mn accumulation in day 12 and day 26 neuroprogenitors after Mn exposure (125 µM). Data is expressed as Mn/DNA (nmol/µg) versus developmental time point. Data are mean ± SEM (n = 3). Statistical significance by one-way ANOVA (*p<0.05).
Mn toolbox is effective in hiPSC-derived mesencephalic floor-plate neuroprogenitors and/or post-mitotic dopaminergic neurons.
(a–i) Activity of selected small molecules at indicated neurodevelopmental stage. Data is expressed as fold change in Mn level of small molecule versus vehicle at indicated developmental time point. Data are mean ± SEM (n = 3). Statistical significance by one-way ANOVA (small molecule versus vehicle) is indicated by white asterisks within the colored bars of the graph and significance by unpaired t-test (day 12 versus day 26) is indicated by asterisks above the brackets between the compared bars; (*p<0.05, **p<0.001).
Discussion
HTS technology permits the rapid testing of thousands of small molecules for their impact on a particular biological system33,34. Our application of HTS is distinct from the more typical HTS approaches targeting specific proteins or enzymatic processes. Our approach would thus be expected to lead to a larger number of potential biological targets, increasing the probability of finding true hits over traditional screens with only one or a few targets. The CFMEA high throughput screen identified a chemically diverse toolbox of 41 small molecules that increase or decrease intracellular Mn levels. Despite testing 40,167 small molecules, only a few chemical structural similarity clusters were identified, suggesting we likely did not identify the complete range of structure-activity relationships that influence cellular Mn status. The vast majority (39/41) of small molecules in the Mn toolbox retained activity at low Mn concentrations, representative of normal brain Mn levels23. This conservation of Mn-modifying activity at normal total Mn levels suggests that the majority of our small molecules target homeostatic processes mediating normal neuronal Mn content, rather than those functioning only under toxicological concentrations.
We have examined potential structure-function relationships by employing a chemical informatics approach to characterize the validated small molecules. Information was found regarding functional targets for five of the 41 small molecules, implicating a major signaling pathway with known links to Mn biology. For example, MAPK14 (p38-α) has been shown to activate in cells responding to cytotoxic levels of Mn, 250 µM Mn or higher35,36. Excessive Mn can increase oxidative stress, COX-2 expression and glutathione depletion37. VU0482585, identified by STITCH analysis (Fig. 4a) as a potential inhibitor of the MAPK14, has confirmed Mn level increasing activity in post-mitotic human dopaminergic neurons (Fig. 4b). Together, these data suggest a potential link between the MAPK14 signaling pathway and control of neuronal Mn content under conditions of excess Mn. The identification of ABCC2 and ABCG2 as putative targets is also noteworthy given their role in unidirectional efflux of toxic substances and known expression in stem cell like populations38,39. Aside from these putative signaling protein targets, our chemical structure associations implicated interactions with enzymes involved in neurotransmission, ACHE and BCHE, consistent with cholinergic disruption occurring with Mn exposure40,41. These and other functional leads suggest our chemical toolbox has high utility for future studies aimed at identifying mechanisms underlying Mn neurobiology.
We tested the hypothesis that mechanisms regulating intracellular Mn content are developmentally regulated in differentiating dopaminergic neurons, given their known susceptibility to Mn toxicity in the context of parkinsonism42,43. This work provides compelling evidence for the developmental regulation of cellular Mn content, as six of the nine small molecules tested showed significant differences in activity between post-mitotic dopamine neurons and floor plate lineage neuroprogenitors (Fig. 6d–i). Furthermore, our data demonstrate the existence of cellular-based Mn handling processes, as pharmacological modes of action acting directly on the Mn ion (e.g. as an ionophore or metal chelator) would not be expected to display developmental stage specific activity. This observation provides strong experimental evidence that modulation of neuronal Mn content is an active cell autonomous process, suggestive that in vivo brain Mn content is not exclusively regulated at the level of the blood-brain and blood-cerebrospinal fluid barriers as indicated by previous studies42,44,45. High evolutionary conservation of Mn handling targets is also strongly suggested by the fact that 9 of 9 tested small molecules identified via a mouse neuronal model showed activity in a human neuronal model system. The fact that the small molecules identified with stage-specific activity included both Mn level increasing and Mn level decreasing small molecules suggests that processes capable of responding to both insufficient and excess Mn are utilized to achieve optimal cellular Mn content.
Given their efficacy in mouse and human in vitro systems, the identified small molecules and their biological targets have potential in vivo applications for manipulating neuronal Mn content. The cellular and behavioral impacts of Mn exposure have been studied in a variety of animal models including fruit flies, nematodes, rodents and non-human primates46,47. Further development of these, or similar, small molecules for in vivo manipulation of neuronal Mn content would enable mechanistic studies into the role of Mn homeostasis in neuronal function and as an environmental risk factor for neurological disease. Pharmacological manipulation of cellular Mn status, coupled with genetic manipulation of the biological targets of these small molecules, would facilitate exploration of Mn-dependent brain functions and complex Mn-biology related behaviors in animal models at a level of control not previously possible.
In summary, we utilized a chemical biology approach to develop a highly diverse small molecule toolbox of neuronal Mn content modifiers. This Mn toolbox is comprised of small molecules capable of modulating intracellular Mn levels under both physiological and toxicological conditions. Biochemical informatics analysis has implicated modulation of cellular signaling pathways with intriguing links to Mn biology and human disease. The identified small molecules exhibit activity in a clinically relevant human dopaminergic neuronal lineage. Furthermore, we report strong evidence for the regulation of intracellular Mn levels over human neurodevelopment. The collection of small molecules described here is the most complete, publically available, resource for interrogation of Mn handling processes and provides a template for development of therapeutic agents for neurological disorders associated with disruption of Mn homeostasis.
Methods
HTS
CFMEA was optimized for HTS to detect both Mn level increasing and decreasing small molecules of neuronal Mn status in mammalian cells. Checkerboard assays, using either 500 µM or 31.25 µM MnCl2 as a positive control relative to the 125 µM MnCl2 used as the baseline Mn exposure condition, confirmed Z’>0.5 in both directions48 and enabling a robust screen in 384-well format (Supplementary Fig. S1). Quality controls were included on each plate for normalization on a plate-by-plate basis. Wild-type StHdhQ7/Q7 immortalized murine striatal cells were plated at a density of 3,000 cells per 384-well 16 hours prior to assay start time. Media was removed and the cells washed by ELX405 Microplate Washer (Bio-Tek) with 1X Hank's-Balanced Salt Solution (HBSS) with calcium and magnesium (Corning) prior to application of 20 µL of 1.5X (15 µM) small molecule in HBSS. Plates were incubated for 30 minutes at 37°C and 10 µL of 3X (375 µM) MnCl2 in HBSS was applied leading to a final concentration of 125 µM MnCl2 and a nominal concentration of 10 µM for the small molecule to be tested. Liquid deliveries were made using a Velocity 11 Bravo liquid handler (Agilent). Plates were incubated for 3 hours at 37°C. Incubation was terminated by a five cycle wash and plate blotting to remove any residual volume. CFMEA was performed by adding 30 µL fura-2 extraction buffer (0.1% Triton-X100, 0.5 µM fura-2 ultra pure (Enzo Life Sciences), 100 µM Dextran Alexa-Fluor 568 (Life Technologies) was loaded by Multidrop Combi Reagent Dispenser (Thermo Scientific). Plates were then incubated for two hours at 37°C and read using FITC filters, 350/25 nm (excitation) and 525/25 nm (emission) for fura-2 and 545/25 nm (excitation) and 620/30 nm (emission) for Alexa 568, by EnVision Multilabel Reader (Perkin Elmer). Data was analyzed with Pipeline Pilot (Acellrys) and Microsoft Excel. Hits were identified in the single point primary screen by z-score and b-score approaches49, then confirmed by duplicate experimental replicates. CRCs were generated by iteratively reweighted least squares, allowing determination of convergence and EC5050,51. Native fluorescence of small molecules was assessed by measurement of 0.5 µM of the test small molecule (or 0.5 µM fura-2 as a positive control) at the same excitation and emission wavelengths used for CFMEA. Fluorescence of each small molecule was compared to +6 S.D. above the DMSO negative control. Cytotoxicity was evaluated by CellTox Green cytotoxicity assay (Promega) under identical conditions as the HTS paradigm. After this period, the CellTox Green dye was added and signal was measured using, monochromatic wavelengths 500 nm (excitation) and 523 nm (emission) using EnSpire Multimode Plate Reader (Perkin Elmer). Toxicity determination included lysed cells as a positive control. Small molecule libraries included collections from NIH Clinical Collection I/II, Microsource Spectrum Collection, kinase inhibitors from GlaxoSmithKline, Enzo Life Sciences and Roche, Vanderbilt non-steroidal anti-inflammatory library (NSAIDs), Vanderbilt bioactive lipid library, ChemBridge and ChemDiv.
Chemical Similarity Analysis
Similarity analysis using the BCL::CHEMINFO Suite was performed on the 41 validated small molecules52. Three dimensional representations of the molecules were generated using Corina53. An upper triangle distance matrix considering largest common substructure between two molecules, (A⋂B)/(A⋃B), was generated using a common subgraph isomorphism algorithm. The vertices and edges were colored by atom type and bond type, respectively. BCL::CLUSTER was used to perform hierarchical agglomerative clustering using complete linkage to generate color-coded dendrograms to visualize structural similarity and activity simultaneously54.
hiPSC neuroprogenitors
A hiPSC line generated from a control subject (CC3), using established methods in our laboratory following the work of Dr. Shinya Yamanaka32,55, was utilized for all human studies. Pluripotency was validated by Pluritest56, euploid karyotype was confirmed (Genetic Associates, Nashville TN) and the absence of the reprogramming plasmid integration was confirmed by PCR.
Mn quantification in human floor-plate lineage dopaminergic neurons
Dopaminergic differentiation of hiPSCs was performed exactly as described31, except LDN193189 (Stemgent) was used at 400 nM. To assess Mn levels on day 12 of differentiation (floor-plate lineage neuroprogenitors) and day 26 of differentiation (post-mitotic dopaminergic neurons) of differentiation, cells were dissociated with Accutase (Stem Cell Technologies) on day 10 or 24 and replated at a density of 50,000 cells/well onto Matrigel-coated 96-well plates containing: 100% N2 medium with ROCK-inhibitor (10 µM, StemCell Technologies) for day 10 cells and Neurobasal medium (Invitrogen) supplemented with B27/glutamax, CHIR (3 µM, Stemgent), brain-derived neurotrophic factor (BDNF) (R&D, 20 ng/mL), glial cell line-derived neurotrophic factor (GDNF) (R&D, 20 ng/mL), ascorbic acid (Sigma, 200 mM), TGFβ3 (R&D, 1 ng/mL), DAPT (Tocris, 10 µM) and dibutyryl cAMP (Sigma, 0.5 mM) containing ROCK-inhibitor (10 µM) for day 24 cells. On day 11 and 25 of differentiation, cells were fed with the aforementioned media without ROCK-inhibitor. Net Mn uptake via the same paradigm utilized in the murine striatal cells (30 minutes pre-incubation with vehicle or small molecule at 15 µM, followed by 3 hours 125 µM MnCl2 in HBSS with continued presence of vehicle or small molecule at 10 µM) was assessed the following day by CFMEA.
Immunofluorescence
Immunocytochemistry of human neural lineages was performed as described55, with the following antibodies: FOXA2 (1:100, BD Biosciences), LMX1A (1:1000, Millipore), β-III tubulin (TU-20) (1:500, Thermo Scientific) and TH (Pel-Freez, 1:500).
STITCH analysis
Protein chemical interactions were predicted with STITCH 4.057. SMILE strings were exported from all validated small molecules with known functional targets. Analysis included structures with Tanimoto scores greater than 0.7. Action view diagrams were generated to illustrate known protein-chemical relationships of all connected nodes.
References
Finney, L. A. & O'Halloran, T. V. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science 300, 931–936 (2003).
Domaille, D. W., Que, E. L. & Chang, C. J. Synthetic fluorescent sensors for studying the cell biology of metals. Nat Chem Biol 4, 168–175 (2008).
Zecca, L., Youdim, M. B., Riederer, P., Connor, J. R. & Crichton, R. R. Iron, brain ageing and neurodegenerative disorders. Nat Rev Neurosci 5, 863–873 (2004).
Choi, B. S. & Zheng, W. Copper transport to the brain by the blood-brain barrier and blood-CSF barrier. Brain Res 1248, 14–21 (2009).
Cousins, R. J., Liuzzi, J. P. & Lichten, L. A. Mammalian zinc transport, trafficking and signals. J Biol Chem 281, 24085–24089 (2006).
Aschner, M., Guilarte, T. R., Schneider, J. S. & Zheng, W. Manganese: recent advances in understanding its transport and neurotoxicity. Toxicol Appl Pharm 221, 131–147 (2007).
Stredrick, D. L. et al. Manganese-induced cytotoxicity in dopamine-producing cells. Neurotoxicology 25, 543–553 (2004).
Josephs, K. A. et al. Neurologic manifestations in welders with pallidal MRI T1 hyperintensity. Neurology 64, 2033–2039 (2005).
Bowler, R. M. et al. Neuropsychological sequelae of exposure to welding fumes in a group of occupationally exposed men. Int J Hyg Envir Heal 206, 517–529 (2003).
Takeda, A. Manganese action in brain function. Brain Res Rev 41, 79–87 (2003).
Aschner, J. & Aschner, M. Nutritional aspects of manganese homeostasis. Mol Aspects Med 26, 353–362 (2005).
Erikson, K., Thompson, K., Aschner, J. & Aschner, M. Manganese neurotoxicity: a focus on the neonate. Pharmacol Therapeut 113, 369–377 (2007).
Bowman, A. B., Kwakye, G. F., Herrero Hernandez, E. & Aschner, M. Role of manganese in neurodegenerative diseases. J Trace Elem Med Bio. 25, 191–203 (2011).
Finkelstein, M. M. & Jerrett, M. A study of the relationships between Parkinson's disease and markers of traffic-derived and environmental manganese air pollution in two Canadian cities. Environ Res 104, 420–432 (2007).
Lucchini, R. G. et al. High prevalence of Parkinsonian disorders associated to manganese exposure in the vicinities of ferroalloy industries. Am J Ind Med 50, 788–800 (2007).
Lechpammer, M. et al. Pathology of inherited manganese transporter deficiency. Ann Neurol 75, 608–612 (2014).
Gonzalez-Cuyar, L. F. et al. Quantitative neuropathology associated with chronic manganese exposure in South African mine workers. Neurotoxicology; 10.1016/j.neuro.2013.12.008 (2013).
Williams, B. B. et al. Altered manganese homeostasis and manganese toxicity in a Huntington's disease striatal cell model are not explained by defects in the iron transport system. Toxicol Sci 117, 169–179 (2010).
Erikson, K. M., Shihabi, Z. K., Aschner, J. L. & Aschner, M. Manganese accumulates in iron-deficient rat brain regions in a heterogeneous fashion and is associated with neurochemical alterations. Biol Trace Elem Res 87, 143–156 (2002).
Rabin, O., Hegedus, L., Bourre, J. M. & Smith, Q. R. Rapid brain uptake of manganese(II) across the blood-brain barrier. J Neurochem 61, 509–517 (1993).
Garrick, M. D. et al. DMT1: a mammalian transporter for multiple metals. Biometals 16, 41–54 (2003).
Tuschl, K., Mills, P. B. & Clayton, P. T. Manganese and the brain. Int Rev Neurobiol 110, 277–312 (2013).
Bowman, A. B. & Aschner, M. Considerations on manganese (Mn) treatments for in vitro studies. Neurotoxicology 41, 141–142 (2014).
Chun, H. S., Lee, H. & Son, J. H. Manganese induces endoplasmic reticulum (ER) stress and activates multiple caspases in nigral dopaminergic neuronal cells, SN4741. Neurosci Lett 316, 5–8 (2001).
Gitler, A. D. et al. Alpha-synuclein is part of a diverse and highly conserved interaction network that includes PARK9 and manganese toxicity. Nat Genet 41, 308–315 (2009).
Kumar, K. K., Aboud, A. A., Patel, D. K., Aschner, M. & Bowman, A. B. Optimization of fluorescence assay of cellular manganese status for high throughput screening. J Biochem Mol Toxic 27, 42–49 (2013).
Kwakye, G. F., Li, D. & Bowman, A. B. Novel high-throughput assay to assess cellular manganese levels in a striatal cell line model of Huntington's disease confirms a deficit in manganese accumulation. Neurotoxicology 32, 630–639 (2011).
Austen, M. & Dohrmann, C. Phenotype-first screening for the identification of novel drug targets. Drug Discov Today 10, 275–282 (2005).
Maggiora, G., Vogt, M., Stumpfe, D. & Bajorath, J. Molecular similarity in medicinal chemistry. J Med Chem 57, 3186–3204 (2014).
Drewry, D. H., Willson, T. M. & Zuercher, W. J. Seeding collaborations to advance kinase science with the GSK Published Kinase Inhibitor Set (PKIS). Curr Top Med Chem 14, 340–342 (2014).
Kriks, S. et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson's disease. Nature 480, 547–551 (2011).
Okita, K., Nakagawa, M., Hyenjong, H., Ichisaka, T. & Yamanaka, S. Generation of mouse induced pluripotent stem cells without viral vectors. Science 322, 949–953 (2008).
Macarron, R. et al. Impact of high-throughput screening in biomedical research. Nat Rev Drug Discov 10, 188–195 (2011).
Inglese, J. et al. High-throughput screening assays for the identification of chemical probes. Nat Chem Biol 3, 466–479 (2007).
Bae, J.-H. et al. Manganese induces inducible nitric oxide synthase (iNOS) expression via activation of both MAP kinase and PI3K/Akt pathways in BV2 microglial cells. Neurosci Lett 398, 151–154 (2006).
Hirata, Y., Furuta, K., Miyazaki, S., Suzuki, M. & Kiuchi, K. Anti-apoptotic and pro-apoptotic effect of NEPP11 on manganese-induced apoptosis and JNK pathway activation in PC12 cells. Brain Res 1021, 241–247 (2004).
Zhang, P., Hatter, A. & Liu, B. Manganese chloride stimulates rat microglia to release hydrogen peroxide. Toxicol Lett 173, 88–100 (2007).
Nies, A. T. & Keppler, D. The apical conjugate efflux pump ABCC2 (MRP2). Pflug Arch Eur J Phys 453, 643–659 (2007).
Patrawala, L. et al. Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and ABCG2- cancer cells are similarly tumorigenic. Cancer Res 65, 6207–6219 (2005).
Finkelstein, Y., Milatovic, D. & Aschner, M. Modulation of cholinergic systems by manganese. Neurotoxicology 28, 1003–1014 (2007).
Santos, D. et al. The inhibitory effect of manganese on acetylcholinesterase activity enhances oxidative stress and neuroinflammation in the rat brain. Toxicology 292, 90–98 (2012).
Olanow, C. W. Manganese-induced parkinsonism and Parkinson's disease. Ann. N. Y. Acad. Sci. 1012, 209–223 (2004).
Kumar, K. K., Aboud, A. A. & Bowman, A. B. The potential of induced pluripotent stem cells as a translational model for neurotoxicological risk. Neurotoxicology 33, 518–529 (2012).
Bornhorst, J. et al. Impact of manganese on and transfer across blood-brain and blood-cerebrospinal fluid barrier in vitro. J. Biol. Chem. 287, 17140–17151 (2012).
Schmitt, C., Strazielle, N., Richaud, P., Bouron, A. & Ghersi-Egea, J.-F. F. Active transport at the blood-CSF barrier contributes to manganese influx into the brain. J. Neurochem. 117, 747–756 (2011).
Au, C., Benedetto, A. & Aschner, M. Manganese transport in eukaryotes: the role of DMT1. Neurotoxicology 29, 569–576 (2008).
Guilarte, T. R. et al. Nigrostriatal dopamine system dysfunction and subtle motor deficits in manganese-exposed non-human primates. Exp. Neurol. 202, 381–390 (2006).
Zhang, J. H., Chung, T. D. & Oldenburg, K. R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen 4, 67–73 (1999).
Brideau, C., Gunter, B., Pikounis, B. & Liaw, A. Improved statistical methods for hit selection in high-throughput screening. J Biomol Screen 8, 634–647 (2003).
Street, J. O., Carroll, R. J. & Ruppert, D. A Note on Computing Robust Regression Estimates Via Iteratively Reweighted Least-Squares. Am Stat 42, 152–154 (1988).
Rousseeuw, P. J. & Verboven, S. Robust estimation in very small samples. Comput Stat Data An 40, 741–758 (2002).
Butkiewicz, M. et al. Benchmarking ligand-based virtual High-Throughput Screening with the PubChem database. Molecules 18, 735–756 (2013).
Sadowski, J. & Gasteiger, J. From atoms and bonds to three-dimensional atomic coordinates: automatic model builders. Chem Rev 93, 2567–2581 (1993).
Alexander, N., Woetzel, N. & Meiler, J. Bcl:: Cluster: A method for clustering biological molecules coupled with visualization in the Pymol Molecular Graphics System. In: IEEE 1st International Conference on Computational Advances in Bio and Medical Sciences, 13–18 (2011).
Neely, M. D. et al. DMH1, a highly selective small molecule BMP inhibitor promotes neurogenesis of hiPSCs: comparison of PAX6 and SOX1 expression during neural induction. ACS Chem Neurosci 3, 482–491 (2012).
Muller, F. J. et al. A bioinformatic assay for pluripotency in human cells. Nat Methods 8, 315–317 (2011).
Kuhn, M. et al. STITCH 4: integration of protein-chemical interactions with user data. Nucleic Acids Res 42, D401–D407 (2014).
Acknowledgements
K.K.K. acknowledges support by the NIGMS T32 GM07347 and the Vanderbilt Medical Scientist Training Program. This work was supported by NIH P30 ES000267 (A.B.B.), R01 ES016931 (A.B.B.), R01 ES010563 and ViCTER RO1 ES010563-13S1 (A.B.B. and M.A.), R01 GM080403 (J.M.), R01 MH090192 (J.M.), R01 GM099842 (J.M.), R01 DK097376 (J.M.), NSF BIO Career 0742762 (J.M.), CHE 1305874 (J.M.) and CI TraCS [OCI-1122919] (E.W.L.). In addition, we thank the members of the Vanderbilt HTS Core: Drs. Paige Vinson and Chris Farmer for their consultation and expertise. We thank Bingying Han, Michael A. Uhouse and John X. Sun for their technical assistance. We recognize GlaxoSmithKline and Roche for access to their kinase inhibitor sets.
Author information
Authors and Affiliations
Contributions
K.K.K., E.W.L., M.D.N., C.D.W., J.M., M.A. and A.B.B. contributed to the writing of the manuscript. K.K.K., E.W.L., A.A.A., M.D.N., J.A.B., C.D.W., J.M., M.A. and A.B.B. participated in the design of the experiments. K.K.K., E.W.L., A.A.A., M.D.N., R.R., J.A.B. and M.O. conducted the experiments. Data analysis was performed by K.K.K., E.W.L., A.A.A., J.M., M.A. and A.B.B.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Kumar, K., Lowe, Jr., E., Aboud, A. et al. Cellular manganese content is developmentally regulated in human dopaminergic neurons. Sci Rep 4, 6801 (2014). https://doi.org/10.1038/srep06801
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep06801
This article is cited by
-
Detection, visualization and quantification of protein complexes in human Alzheimer’s disease brains using proximity ligation assay
Scientific Reports (2023)
-
HrpA anchors meningococci to the dynein motor and affects the balance between apoptosis and pyroptosis
Journal of Biomedical Science (2022)
-
Genomic, transcriptomic, and metabolomic profiles of hiPSC-derived dopamine neurons from clinically discordant brothers with identical PRKN deletions
npj Parkinson's Disease (2022)
-
Influence of Substrate Stiffness on Barrier Function in an iPSC-Derived In Vitro Blood-Brain Barrier Model
Cellular and Molecular Bioengineering (2022)
-
Relevance of biometals during neuronal differentiation and myelination: in vitro and in vivo studies
BioMetals (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.