Abstract
Dendritic trees influence synaptic integration and neuronal excitability, yet appear to develop in rather arbitrary patterns. Using electron microscopy and serial reconstructions, we analyzed the dendritic trees of four morphologically distinct neocortical interneuron subtypes to reveal two underlying organizational principles common to all. First, cross-sectional areas at any given point within a dendrite were proportional to the summed length of all dendritic segments distal to that point. Consistent with this observation, total cross-sectional area was almost perfectly conserved at bifurcation points. Second, dendritic cross-sections became progressively more elliptical at more proximal, larger diameter, dendritic locations. Finally, computer simulations revealed that these conserved morphological features limit distance dependent filtering of somatic EPSPs and facilitate distribution of somatic depolarization into all dendritic compartments. Because these features were shared by all interneurons studied, they may represent common organizational principles underlying the otherwise diverse morphology of dendritic trees.
Similar content being viewed by others
Introduction
The morphologies of dendrites directly influence synaptic integration and neuronal excitability. The passive properties of dendrites can present barriers to effective propagation of voltage signals from distal synaptic locations to the soma and axon, where action potentials are generally initiated. The intrinsic conductance and capacitance of dendritic membrane attenuates and broadens synaptic potentials in a distance-dependent manner that is exacerbated by the asymmetry of tapering dendrites1,2,3,4,5.
However, tapering dendrites also have progressively decreasing diameters and surface areas, which will increase the local input impedance and reduce local capacitance at more distal dendritic locations1,2. By generating fast and large local synaptic responses at distal locations, tapering dendrites limit the functional impact of voltage attenuation in dendritic trees, a process sometimes referred to as “passive normalization,”6 and optimize the conduction of synaptic currents within dendrites7. The structure of dendritic trees also influences the excitability of neurons and the pattern of action potential output8,9, even when dendritic trees have the same total dendritic length and surface area10,11. By influencing the transfer of current between soma and dendrites, the geometry of dendritic branch points can regulate the impact of variable dendritic topology on synaptic integration and excitability10.
Dendritic trees come in many shapes and sizes. Although similarities in dendritic morphology is often used to classify neurons into discrete subtypes, no two dendritic trees are exactly alike: individual dendritic segments appear tortuous and rather arbitrary in the locations of branch points. We asked whether there are common principles underlying the morphologies of dendritic trees in cortical GABAergic interneurons. Because light microscopy lacks the resolution to accurately measure dendritic dimensions at the sub µm level, we combined light microscopy and reconstructions of serial electron microscopy to compare dendritic dimension and shape in four neocortical nonpyramidal cell subtypes12,13,14,15,16. Our data reveal several conserved morphological characteristics of dendritic trees common to all subtypes examined that limit distance dependent filtering of somatic EPSPs and facilitate distribution of somatic depolarization into dendritic compartments. These features may represent common organizational principles underlying the otherwise diverse morphology of dendritic trees.
Results
Neurolucida reconstructions of biocytin-filled nonpyramidal neurons from slice preparations of rat frontal cortex15,16 were used to analyze the arborization of dendritic trees from a Martinotti (MA) cell, a fast-spiking basket cell (FS), a double-bouquet (DB) cell and a large basket (LB) cell ( Fig. 1a ). Physiological properties of these neurons are shown in Supplementary Table 1. Dendritic arborizations were reconstructed three dimensionally by Neurolucida (reconstructions available at NeuroMorpho.Org, http://neuromorpho.org/neuroMorpho/index.jsp). These cells were selected because of their mostly intact dendritic trees ( Fig. 1b ) within the 300 µm thick slices of tissue. Eighty-five dendritic segments (red circles on dendrograms in Fig. 1b , mean length 16.8 ± 6.8 µm) were selected at ∼50 µm intervals along dendritic branches lacking severed endings. Some dendritic segments included bifurcation points. Dendritic segments were serially sectioned, observed under electron microscopy (EM) and reconstructed into 3D morphologies ( Fig. 1c, d , see Supplementary Fig. 1 online).
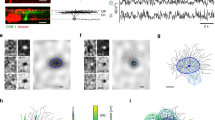
Dendritic structures of neocortical cells representing 4 nonpyramidal neuron subtypes.
(a) Morphology of the neuron. Axons are shown in red, dendrites and somata are shown in black. The MA cell extends its axonal arbors into layer I, whereas the FS cell has a horizontally elongated axonal arborization. The DB cell has a descending narrow axonal arborization and the LB cell has a large soma and basket terminals. (b) Dendrograms of the same neurons. Red circles indicate segments processed for 3D reconstruction from successive EM sections. Asterisks indicate cut ends of dendrites. Dendritic trees are labeled (M1-M4, F1-F9, D1-D5, L1-L5) and referenced in later figures. Ultrastructure images for segment p, m, d of Martinotti cell are shown in Figure 3 g–i, respectively. (c) Ultrastructure showing dendrite (Den) and spine head (Sp) of the MA cell, which correspond with the dendritic segment indicated by arrow head in (a) and (b). White arrows indicate synapses occurring on those structures. (d) Multi-focus photograph of the dendrites indicated by arrowheads in (a) and (b), along with reconstructed dendrites made from successive EM images of the same dendrite segments. Gray arrow indicates the spine and dendrite shown in (c).
To explore how dendrites tapers with distance, we first investigated the relationship between dendrite cross-sectional area and distance from the soma ( Fig. 2a , upper panels). Cross sectional areas were calculated by dividing the volume of a dendritic cylinder by its length (0.18 – 2.7 µm), where no spines were attached, to generate an average value for the cylinder. The average circumference of the dendrite was calculated by dividing the surface area of the dendritic cylinder by its length. These methods reduced potential variability when choosing 2-dimensional cross-sectional planes. Proximal dendritic sections varied in cross-sectional areas (0.1 – 2.2 µm2)17, while all distal dendritic segments had small cross-sectional areas (∼ 0.5 µm2). Dendritic thickness steeply tapered over the first 50 µm of dendrite, where bifurcations were frequently found ( Fig. 1b , 2a ). Toward the distal end of dendritic branches, branching was infrequent and tapering became less extreme, but remained significant. Trajectory lines drawn between sequential dendritic segments ( Fig. 2a , lower panels) revealed that parent dendrites bifurcated into two smaller daughter branches of different sizes. Smaller branches occurring after initial bifurcations at proximal locations fully accounted for all of the small cross-sectional dendrites located near the soma. These thin proximal dendrites extended for only short distances and typically ended without additional bifurcations, whereas thicker proximal daughter dendrites extended for long distances and had frequent additional bifurcations. While dendrite cross sectional areas varied across cells and within individual cells, remarkably, for any dendritic location, the cross-sectional area was well correlated with the total dendritic length expanding distally from the point of reference ( Fig. 2b , see also Supplementary Fig. 2 online). For this analysis, we used only reconstructed segments having natural, non-severed endings.
Dendritic dimensions of nonpyramidal cell subtypes.
(a) upper panel: The cross-sectional areas of dendritic segments plotted against their distances from the soma. Large dendrites are frequently found near somata, whereas only small dendrites are observed at distal locations. Some small dendrites were also found in proximal locations. lower panel: cross-sectional areas of dendritic segments shown with trajectory lines for some trees (M2, M3, F4 – F6, D1, D2, L2 – L4). At the dendritic bifurcations, the daughter branches split into larger (black circle) and smaller (white circle) segments. (b) For all nonpyramidal cells tested, dendritic cross-sectional areas were well correlated with the total dendritic length distal from the point of reference.
Our reconstructions revealed that dendritic cross-sections do not approximate circles, but rather are irregular ellipses. We quantified this feature using an “ellipticity index” determined by dividing the cross-sectional area of the reconstructed dendrite by the area of a true circle having the same circumference, where the index of true circle is 1 and that of line is 0. Across all neurons, dendritic cross-sections had a mean ellipticity index of 0.64 ± 0.15 (range: 0.33 – 0.95, n = 163) ( Fig. 3 ). FS cell dendrites were slightly, but significantly, flatter than those of double bouquet cell (p = 0.029, Fig. 3c , Table 1 ). In dendrites from all neurons, smaller and more distal dendrites tended to be more circular than more proximal, larger dendrites ( Fig. 3a ). Interestingly, we found a linear relationship between cross sectional area and circumference in all neurons studied ( Fig. 3e ), suggesting the dendritic volume and surface area decrease at a constant rate with distance from the soma, which would not be true for dendrites with circular cross-sections. As shown in Figure 3f , the surface area to volume ratio is relatively uniform at dendritic locations more distal than 30–40 µm from the soma. At more proximal locations, surface area to volume becomes smaller. However proximal dendritic segments tend to be filled with greater amounts of intracellular membrane-bound compartments ( Fig. 3g–i ; e.g., mitochondria, endoplasmic reticulum, Golgi apparatuses, etc.), suggesting that our measurements may disproportionately underestimate surface area to cytoplasmic volume ratios in these proximal locations.
The cross-sections of nonpyramidal cell dendrites are irregular ellipses.
(a) Ellipticity Index plotted against cross-sectional area for dendritic segments in each of the four neurons studied. Ellipticity indices were generally between 0.4 and 0.9 in most dendrites. Proximal (thicker) dendrites tended to be flatter than distal (thinner) dendrites. The index of a circle is 1 and that of a line is 0. (b) Reconstructed image of a dendrite segment of the Martinotti cell. The cross-section is an ellipse. (c) Average ellipticity index of the FS cell is smaller than those of the double bouquet cell (*Tukey, p = 0.029). MA, Martinotti cell; FS, FS basket cell; DB, double bouquet cell; LB, large basket cell. (d) Rotating 3D image of the elliptical dendritic segment of the large basket cell. The apparent thickness varies greatly when viewed from different angles (arrows) and pronounced diameter fluctuations are apparent along the segment length when viewed from a single point of view, as would occur under light microscopy. (e) Cross-sectional areas plotted against perimeter of the cross-section showing a strong linear correlation between the two variables. Line of true circle (red doted curve) and liner fit line are shown. The larger dendrites are located below the curve of true circle, although the smaller dendrites are tend to be on or quite near the curve. It suggested the smaller dendrites tended to be true circle, but the larger ones were rather elliptical in their cross section. (f) Surface-area-to-volume ratios plotted against distance from soma showing constant especially from 30 – 40 µm from soma or more distal. (g–i) Ultrastructure of Martinotti cell dendrites shown as p, m, d in dendrogram of Figure 1b . Note that the proximal dendrite (g) contains many organelles, while the middle (h) and distal dendrites (i) contain only a small number of organelles. Scale bar, 1 µm.
Our reconstructions also revealed that dendrites twist as they extend outward, a feature that may account for the perceived diameter fluctuations of dendrites when they are observed under light microscopy ( Fig. 3d and see Supplementary Video 1 online). For this reason, measurements of dendritic diameters using light microscopes may not be reliable18.
In order to facilitate computational modeling of voltage signals in dendrites, Rall described a special case in which dendritic branches have uniform input conductance between parent and daughter dendrites, Rall’s so called “three-halves power law,” which allows dendritic trees with uniform electrotonic distances at terminal branches to be modeled as simple cables3,19,20. While some attempts have been made to measure the diameter of parent and daughter branches using light microscopy, it is still controversial whether branch points in real neurons follow precise rules, such as Rall’s 3/2 power law21,22,23,24. We analyzed EM 3D-reconstructions of dendritic branch points ( Fig. 4a ) to directly address this issue and found that the cross-sectional area of parent dendrites matched well with the summed cross-sectional areas of the two daughter branches ( Fig. 4b ). To quantify this relationship, we calculated a cross-sectional area “matching index” by dividing the summed dendritic cross-sectional area of the two daughter branches by the cross-sectional area of the parent dendrite ( Table 2 ). We found that the majority of branches in all cell types had near-perfect matching of parent-to-daughter cross-sectional areas. Matching cross-sectional areas might allow a constant number of microtubules to transit through bifurcation points toward distal dendrites ( Fig. 4c ), which could optimize transport mechanisms in dendritic trees. Indeed, using serial SEM images having 7 or 10 nm z steps, we found that the number of microtubules in dendritic segments is linearly correlated with their cross-sectional areas ( Fig. 5a–f ). We also counted the numbers of microtubules in parent and daughter branches in two weakly stained calretinin-positive (characteristic of GABAergic double-bouquet cells) bifurcation points treated with nickel DAB. The number of microtubules in the parent branches (60 and 62 microtubules) were almost identical to the total number of microtubules in daughter branches (62 and 63 microtubules, respectively). We also traced microtubules through unlabeled dendritic bifurcations (n = 4; probable pyramidal cells) using the serial SEM images ( Fig. 5g–i , Supplementary Video 2 ). The microtubules in the mother dendrite were found to run continuously into one of the two daughter dendrites ( Fig. 5k, l , Supplementary Video 3 ). Thus the total number of microtubules was almost identical in parent and daughter branches. These results suggest that the conservation of cross-sectional area at branch points may function to maintain a consistent number of microtubules in parent and daughter branches to facilitate microtubule-dependent transport into dendritic trees.
Microtubule distribution at dendritic branch points.
(a) The number of microtubules is linearly correlated with dendritic cross sectional areas. Black circles indicate spiny dendrites and white circles indicate aspiny dendrites. (b – f) Serial FIB/SEM images of a dendrite (10 nm pitch, 5 nm lateral resolution). The same microtubules can be traced from section to section easily. Scale bar, 0.5 µm. (g – j) Serial FIB/SEM images of a bifurcated dendrite (300 nm pitch, 5 nm lateral resolution). The mother dendrite (g) gradually changes shape (h, i) and bifurcates into two daughter dendrites (arrows, (j)). Scale bar, 1 µm. k, A 3D reconstruction of the bifurcated dendrite shown in (g – j). (l) Traces of the microtubules observed in the bifurcated dendrite shown in (k). Microtubules present in the mother dendrite run into one of the two daughter dendrites
Rall‘s computational simplification of bifurcation points is known as the “3/2 power law.”3 We evaluated conductance-matching at dendritic bifurcations in the nonpyramidal neurons using the formula

where c is the circumference and a is the cross-sectional area of the dendrite, with p indicating parent dendrite and d1 and d2 being daughter dendrites (see Methods). We found that the majority of branches had near-perfect matching of parent-to-daughter  , which, at least for cortical interneurons, substantiates the original assumptions Rall used for simplification in his computational models (
Fig. 4d
). To quantify this relationship, we calculated the conductance-matching index by dividing the summed dendritic
, which, at least for cortical interneurons, substantiates the original assumptions Rall used for simplification in his computational models (
Fig. 4d
). To quantify this relationship, we calculated the conductance-matching index by dividing the summed dendritic  of the two daughter branches by the
of the two daughter branches by the  of the parent dendrite (
Table 2
). Matching the conductance of parent and daughter branches may compensate for the impact of variable dendritic topology on synaptic integration and spike generation10.
of the parent dendrite (
Table 2
). Matching the conductance of parent and daughter branches may compensate for the impact of variable dendritic topology on synaptic integration and spike generation10.
We were surprised that branch points in cortical nonpyramidal neurons showed near-perfect matching of parent-to-daughter cross-sectional areas as well as conductance-matching. To compare the two relationships, we plotted the two formulas


on a single graph to identify those values that satisfy both formulas simultaneously. To plot both relationships, we transformed equation (3) into a form using E (ellipticity index) and a (cross sectional area) (see Methods),

Using our measurements for the cross-sectional areas of the parent dendrites, we plotted the range of possible solutions that allow for matching of cross-sectional areas and conductance for daughter dendrites. We found that for the dimensions typical of dendrites, these two relationships are quite similar to each other and that values for daughter dendrites consistently fell near the predicted ranges for both relationships ( Fig. 4e , Table 1 ). We measured the minimum distance between data obtained from the 3D reconstructed daughter branches and compared them with the values predicted for matching of cross-sectional area and conductance. We found that dendritic branching in the large basket cell fell closer to the expected values calculated for cross-sectional area matching, but that data for other cell types did not show significant preference for either concept (p > 0.05, Fig. 4e ). Thus, branch points of cortical nonpyramidal cells tend to conserve both cross-sectional area and conductance-matching.
To test whether there might be a functional significance of having dendritic bifurcations that follow Rall’s “3/2 power law,”3 we simulated synaptic input in computational models of reconstructed neuron morphologies (“real morphology” models; available at ModelDB: https://senselab.med.yale.edu/modeldb, model number 140299) and morphologies modified to eliminate branch point conductance-matching (“conductance-mismatch” models) ( Supplementary Fig. 3 and see Methods). We stimulated dendrites with a synaptic current (alpha function, 2 nS) at different locations in the dendritic tree (10 µm intervals). In morphologically realistic model neurons, the peak somatic depolarization resulting from excitatory post synaptic potentials (EPSP) ( Fig. 6a ) showed limited synapse location-dependent variability (black line near bottom in Fig. 6c and colored lines in Fig. 6d ), while local EPSPs at the site of synaptic input ( Fig. 6b ) were highly variable and location-dependent (colored lines in Fig. 6c ). In conductance-mismatch models, in which branch points do not show conductance-matching, there was larger variability in peak somatic EPSP amplitudes (black line near bottom in Fig. 6e and colored lines in Fig. 6f ) as quantified by the coefficients of variation for somatic EPSP amplitudes ( Fig. 6g ). We next examined passive invasion of steady-state somatic depolarization into dendritic trees. Somatic current injection (40 pA) provided a more uniform depolarization of dendritic branches in neurons having morphologically authentic branch points, with less variation in peak depolarization across locations in the dendritic tree ( Fig. 7a ). In contrast, somatic current injection generated variable depolarization of dendrites in the conductance mismatch models ( Fig. 7b ), with the magnitude of variability increasing with distance from the soma ( Fig. 7c ).
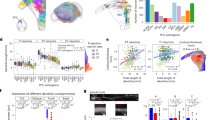
Functional significance of branch point geometry.
(a–f) Attenuation of synaptic responses in dendrites. (a, b) Somatic (a) and local (b) responses to synaptic currents generated in different parts of the dendritic trees in “real morphology” model cells. Inset shows the injected alpha function synapse current (2 nS). (c) Plots of peak of somatic (black) and local synaptic depolarization (color-coded as dendrogram shown in h) in response to synaptic input to various parts of the dendrite of the real morphology model cells. (d) Peak somatic depolarization as shown in (c) but at a larger scale. (e) Plots of peak depolarization at the soma (black) and at the site of synaptic input (color-coded as in h) for synapses occurring in the dendrites of the “conductance-mismatch” models. (f) Peak somatic depolarization as shown in (e) but at a larger scale. Deviations among different branches are larger than the real model cells (d). (g) Plots of the coefficients of variation (CV) of peak somatic depolarization in response to synaptic input to dendritic compartments at a given distance from the soma for the models exhibited in (d) and (f). CVs in the real morphology model cell (blue circles) are very low across the entire dendritic length, while CVs in the conductance mismatch model cell (red circles) increase with distance from soma. (h) Color-coded dendrogram of the model cells used in simulations.
Dendritic branch points are optimized for uniform distribution of somatic voltage into the dendritic trees.
(a), Plot of peak depolarization of dendritic compartments in response to a 40 pA somatic steady current pulse injection into the “real morphology” model cells. Dendritic depolarization is fairly evenly distributed in dendritic branches. Primary dendrites and their associated trees, are color-coded as in Fig. 6h . (b) Plot of peak depolarization of dendritic compartments in response to a 40 pA somatic steady current pulse injected into the soma of neurons having realistic morphologies. Color-coding as in Fig. 6h . (c), Plot of the coefficient of variation (CV) of peak depolarization for all dendritic compartments of a given distance from the soma (vs. distance from the soma) for the models shown in (a) and (b). CV of dendritic depolarization for the real morphology model cell (blue circle) is very low for entire dendritic length, On the other hand, the CV of dendritic depolarization in the conductance mismatch model cell (red circle) increases with distance from soma. (d) Surface areas of dendrites plotted against the surface areas of the soma as measured in the real morphology model cells. (e) The volumes of dendrites are plotted against the volumes of somata as measured in the real morphology model cells
Lastly, we investigated the relationship between somatic size and dendritic trees in the morphologically realistic models. We found a linear relationship between somatic and dendritic volume and between somatic and dendritic surface area among the four neuron subtypes ( Fig. 7d, e , Table 3 ). Somatic volumes were about 1.4 times larger than total dendritic volumes, while somatic surface areas were about 7% of total dendritic surface areas. These data are consistent with the hypothesis that larger somata, containing more intracellular organelles such as ribosomes, may be required to support larger and more elaborate dendritic trees.
Discussion
Our results reveal several features of dendritic structure that underlie the otherwise diverse and variable morphologies of dendritic trees. First, dendritic cross-sectional areas were found to be proportional to the total lengths of all distal dendritic segments, regardless of the number of distal branch points. Second, nonpyramidal neuron dendrites were found to be elliptical, rather than circular, with the degree of ellipticity decreasing with dendritic size and increasing with distance from the soma. This feature tends to limit the variability of surface-area to volume ratios along dendrites. Finally, we found branch points conserve the total cross-sectional area of the parent dendrite. The close match between parent and daughter dendrite cross-sectional area may optimize microtubule-dependent transport within dendrites and may promote efficient electrical distribution of somatic depolarization into dendritic branches, a process that can reduce the influence of dendritic topology on neuron excitability10,11. This morphological property also optimized the fidelity of EPSP propagation to the soma. In addition, we found that, across all neuron subtypes, somatic volume and surface areas are predictive of total dendritic volumes and surface areas. While our results were obtained from the neurons of adolescent rats, we found similar results in immunohistochemically stained dendrites of nonpyramidal cells in the cortex of adult rats fixed in vivo (22 segments, data not shown, see Immunohistochemistry in Methods.).
The linear relationship between the cross-sectional area and the total dendritic length of all dendrites distal to that point indicates that dendrites taper smoothly, independent of branch points, a morphological feature that may facilitate efficient intracellular transport into dendritic trees by allowing an initially large number of microtubules near the base of the dendrite to course continuously from the soma to all distal dendritic endpoints. The conservation of total cross-sectional area at branch points is consistent with this hypothesis in allowing even distribution of trafficked molecules into the dendritic tree. Indeed, in two interneuron branch points examined for microtubule content, we found that the total number of microtubules in daughter branches was almost identical to the number found in the parent dendrite.
The irregular elliptic shape of dendritic cross-sections may provide more efficient packing of neuropil within small volumes of space, especially given the presence of fibers coursing in all directions (vertically, horizontally and radially) through the tissue. Additional studies of neuron packing efficiency are needed to determine if elliptic dendritic segments are indeed advantageous for neuropil organization. Alternatively, elliptical cross-sections at proximal locations will tend to preserve surface area to volume ratios along dendrites. Indeed, we found that dendrites have a fairly constant surface area to volume ratio at locations more distal than 30–40 µm from soma. This may benefit neurons by normalizing gas exchange and molecular diffusion into compartments having different diameters. It would also tend to normalize cytosolic ion concentrations following activation of ion channels uniformly distributed in the dendritic membrane. For instance, it could allow given density of calcium channels to produce a standardized intracellular calcium concentration, independent of dendritic location, to preserve the fidelity of Ca-protein interactions25.
Changes in dendritic morphology are well documented in neurodegenerative diseases. Our data suggest that, in healthy neurons, dendritic structure is more precisely regulated than might be guessed given the diversity of dendritic tree morphologies. It will be important for future work to assess the detailed morphology of dendrites in pathological tissue to test if alterations in dendritic tapering and branch point uniformity might participate in generating the cognitive deficits associated with disease.
Together, our data from four different nonpyramidal cell subtypes reveal morphological features of dendritic trees that appear well suited to allow the uniform retrograde distribution of molecules and electrical signals into dendritic trees, while at the same time, enhancing the anterograde synaptic signal conduction and limiting the effect of dendritic topology on action potential output. Because these features are conserved across cortical nonpyramidal cell subtypes that differ greatly in sizes, shape and functions within cortical circuits, it is likely that these features of dendrites are common to the dendrites of many neuron types throughout the brain.
Methods
Slice recording
Sections of frontal cortex of young Wistar rats (19–23 days postnatal) were cut to a thickness of 300 µm in oblique horizontal cutting plane and immersed in a buffered solution containing (in mM) NaCl, 124.0; KCl, 3.0; CaCl2, 2.4; MgCl2, 1.2; NaHCO3, 26.0; NaH2PO4, 1.0; and glucose, 10.0, aerated with a mixture of 95% O2 and 5% CO2. Cortical cells were targeted for whole-cell recording on a fixed-stage at 30–31°C using a 40x water immersion objective. The electrode solution consisted of (in mM): potassium methylsulfate, 120; KCl, 5.0; EGTA, 0.5; MgCl2, 1.7; Na2ATP, 4.0; NaGTP, 0.3; HEPES, 8.5; and biocytin, 17.
Histology
Tissue slices were fixed in 4% paraformaldehyde, 1.25 or 0.05% glutaraldehyde and 0.2% picric acid in a 0.1 M phosphate-buffered (PB) solution and resectioned into 50 µm thick slices. Slices were incubated with avidin-biotin-peroxidase complex (VECTASTAIN Elite ABC Kit PK-6100, Vector Laboratories, Burlingame, U.S.A.), in Tris-HCl buffered saline (TBS) with or without 0.04% Triton X-100 (TX) and reacted with 3,3-diaminobenzidine tetrahydrochloride (DAB) (0.05%) and H2O2 (0.003%). Slices were then postfixed in 1% OsO4 in PB, dehydrated and flat embedded on silicon-coated glass slides in Epon. The osmium/epon fixation process generates < 10% distortion in x, y and z directions16. The shrinkage was not corrected in the analysis. They were reconstructed with Neurolucida (MicroBrightField, Williston, VT) with a 60x objective lens. We selected 4 interneurons representing each of 4 nonpyramidal cell subtypes: a regular spiking nonpyramidal (RSNP) MA cell, a fast spiking FS cell, an RSNP DB cell and a BSNP LB cell. Neurons were selected based on the intactness of their dendritic trees, having only minimal dendritic severing during slice preparation ( Fig. 1b ). The stained cells were photographed at 0.5 µm focus steps using a 100x objective and dendritic segments identified for EM observation were marked on those photo images. Each subtype had the stereotypical morphological properties of axonal and dendritic arborizations previously associated with those subtypes15,16. From these neurons we selected 14 to 29 dendritic segments to make 3D reconstructions using Visilog software (Noesis, Vélizy, France). Segments without bifurcations were chosen every 50 µm along dendrites lacking artificially cut ends. We also chose bifurcation points from all subtypes to reconstruct. Cells were then serially sectioned into 90 nm thick slices using an ultramicrotome (Reichert Ultracut S, Leica Microsystems, Wetzlar, Germany). Ultrathin serial sections mounted on formvar-coated single-slot grids (NOTCH-NUM Grids, 1 × 2 mm slot, SynapTek) were stained with lead citrate. The segments for EM observation were identified by comparison of EM images and light microscopic images. Electron micrographs were taken with a Hitachi H-7000 electron microscope, using tilting of up to 60°. The thickness of ultra thin sections was calibrated by a color laser 3D profile microscope (VK-9500; Keyence, Japan)26. EM images of the segments and associated structures were captured using a CCD camera (Kodak Megaplus 1.4i, Princeton Instruments, USA) and reconstructed using a 3-D reconstruction system with the software developed by Noesis (Vélizy, France) as an extension of their Visilog program. The mean length of the segments was 16–18 µm (minimum 3.7 µm, maximum 34.3 µm) and 7 – 13% of the total length of the nonpyramidal cell’s dendrites was reconstructed 3 dimensionally (Table 1). These segments were reconstructed from 35 – 145 successive ultrathin sections (90 nm thickness). Dendritic cross-sectional area and circumference were measured as averaged value of a short part of the reconstructed dendrite segment, cross-sectional area was determined as volume divided by length of a portion of the segment and circumference was defined as surface area divided by length of the same portion ( Fig. 4a ). The average length of each segment portion was 724.1 ± 407.1 nm (range: 183.1 – 2737 nm, n = 164).
Immunohistochemistry
Three male Wistar rats (6 weeks of age, 140–160 g) were anesthetized with an overdose of Nembutal and perfused through the heart with 10 ml of a solution of 250 mM sucrose, 5 mM MgCl2 in 0.02 M phosphate buffer (pH 7.4) (PB), followed by 300 ml of 4% paraformaldehyde containing 0.2% picric acid and 0.1% glutaraldehyde in 0.1 M PB. Brains were then removed and oblique horizontal sections (50 µm thick) of frontal cortex were cut on a vibrating microtome (Leica VT1200S, Nussloch, Germany). Tissue sections were dehydrated in 10% sucrose in PB, followed by 25% sucrose and 10% glycerol in PB for 2 hours, frozen in liquid nitrogen. The sections were then incubated in 0.1 M PB containing 1% sodium borohydrate for 30 minutes and in 0.05 M Tris-buffered saline (TBS) containing 1% H2O2 for 30 minutes before incubation with primary antiserum against either somatostatin developed in rabbit (generous gifts from Dr. R Benoit, mixture of S309 [1:1000] and S320 [1:4000]), parvalbumin developed in mouse (1:4000, P-3171, Sigma-Aldrich, Saint Louis, MO, U.S.A.), or calretinin developed in mouse (monoclonal; Chemicon, Temecula, CA, U.S.A., MAB1568; 1:2500) diluted in TBS containing 10% normal goat serum and 2% bovine serum albumin overnight at 4°C. The sections were then incubated in biotin-conjugated secondary antiserum (1:200, BA-1000 or BA-2000, Vector Laboratories) followed by the ABC (Vector Laboratories) and staining with DAB with nickel (0.02% DAB, 0.3% nickel in 0.05M Tris HCl buffer). The stained sections were post-fixed for 40 minutes in 1.5% potassium ferrocyanide and 1% osmium tetroxide, followed by 1 hour in 1% osmium tetroxide alone and dehydrated in graded dilutions of ethanol with 1% uranyl acetate added at the 70% ethanol dehydration state. After flat-embedding in Epon, the sections were observed and photographed under the light microscope as described under ‘Histology’. Then slices were serially re-sectioned at a thickness of 90 nm using the ultramicrotome. The labeled dendrites and associated structures were photographed and digitized from EM negatives using a scanner (GT-9800F, Epson, Suwa, Japan). The structures were reconstructed using the 3D reconstruction software, Reconstruct; http://synapses.clm.utexas.edu/tools/index.stm.
FIB/SEM image capture (Microtubule analysis)
Two male Wistar rats (8 weeks of age, 240g) were anesthetized with an overdose of Nembutal and perfused through the heart with 10 ml of a solution of 250 mM sucrose, 5 mM MgCl2 in 0.02 M phosphate buffer (pH 7.4) (PB), followed by 300 ml of 2% paraformaldehyde containing 0.2% picric acid and 2% glutaraldehyde in 0.1 M PB at 37°C. Warming up the perfusion solution to body temperature preserves the structure of microtubules. Brains were then removed and oblique horizontal sections (50 µm thick) of frontal cortex were cut on a vibrating microtome (Leica VT1200S, Nussloch, Germany). The sections were post-fixed for 40 minutes in 1.5% potassium ferrocyanide and 1% osmium tetroxide, followed by 1 hour in 1% osmium tetroxide alone and dehydrated in graded dilutions of ethanol with 1% uranyl acetate added at the 70% ethanol dehydration state. Tissue was then flat-embedded in Epon.
For electron microscopy observation, the Epon blocks containing cortical layer II/III were glued to an aluminium stub using silver paint. Besides the top surface, the whole block was covered by silver paint to avoid charging the epoxy. The top surface to be studied was coated by several 10 nm thick layers of iridium using a sputter coater. For 3D analysis the mounted blocks were transferred in a combined FIB/SEM (Focused Ion Beam / Scanning Electron Microscope) system using the Carl Zeiss Auriga40 Crossbeam® (Carl Zeiss NTS GmbH, Oberkochen, Germany). In a combined FIB/SEM system, very site specific regions of interest near the surface of embedded tissue in epoxy can be directly cross-sectioned and imaged for three dimensional analysis. The z-resolution is given by FIB slice thickness which can be in the nanometer scale. Regions of interest were identified by SEM imaging the top surface of the block and by comparison with light microscopy images. The top of the regions was protected by ion beam induced deposition of platinum (25 um*25 um area, 1 nA ion beam current, 900 sec deposition time). A view channel for SEM observation was coarse milled and the region was separated from the bulk by two side cuts using a 16 nA ion beam. With a 2 nA ion beam, the first coarse cross-sectioning was medium polished. Using a 600 pA ion beam probe, the surface of the cross-section was further fine polished and slices of 10 nm thickness were milled. After each slice, the milling was paused and the freshly exposed surface of the cross-section was imaged at 1.5 kV acceleration potential using the in-column energy selective back-scattered electron (EsB) detector. The retarding potential of the EsB detector was set to 1314 V (data cube A) or 1014 V (data cube B). By low kV imaging and detection of the low loss back-scattered electrons of the stained tissue surface, imaging was achieved. Using contrast inversion, TEM-like contrast and comparable imaging information was achieved. The acquired images have a 2048 * 1536 store resolution and a image pixel size of 5 nm (data cube A) or 7 nm (data cube B). The cross-section was imaged by the SEM at 54 deg stage tilt and the tilt of the cross-section vs. the electron beam was corrected (tilt angle of 36 deg was used for correction). The milling time per slice was between 15.8 and 17.5 sec. The imaging time per slice was about 52 sec. The serial image alignment and tracing the microtubules were done using Fiji image processing package: http://fiji.sc/wiki/index.php/Fiji, plugins “Register Virtual Stack” or “TurboReg” for the alignment and “TrakEM2” for the tracing. The structures were reconstructed using the 3D reconstruction software, Reconstruct (available at http://synapses.clm.utexas.edu/tools/index.stm)
Statistics
We used Mann Whitney U test for statistical analysis and ANOVA (followed by Tukey test) for ellipticity index of 4 subtypes of nonpyramidal cells for Fig. 3c and Table 1 and Wilcoxon signed-rank test (non-parametrics) for Fig. 4e .
Rall’s model and ramified part of 3D reconstructed dendrite
1) Rall’s model equation is transformed to simple equation using circumference and cross-sectional area.

Rall’s conductance-matching assumption
Where GN (p) is input conductance of parent dendrite, GN (d) is input conductance of daughter dendrite. Conductance is determined by

Where Ri is the specific intracellular resistivity (Ω-cm), Rm is the specific membrane resistivity (Ω-cm2) and r is the radius of dendrite. The Rall model (equation (5)) can be defined by the following equation:

We remove common constants  from both side of the equation and obtain the following:
from both side of the equation and obtain the following:

c is the circumference of dendrite and given by  .
.
a is the cross-sectional area of dendrite and given by  .
.
Putting these together,

The square root of equation (9) is

It is convenient to divide this equation by 

Equation (8) is transformed by equation (11)

We remove common constants  from both side of the equation (12) and obtain the following:
from both side of the equation (12) and obtain the following:

2) The equation (8) is transformed to equation using cross-sectional area and ellipticity index E.
We have a simple relation between radius r and cross section area a using ellipticity index E as follows:

Then the equation (8) is transformed just by substituting  as follows:
as follows:

Simulation analysis
Simulations were done using NEURON 7.127. The “real morphology” models faithfully represented our morphological results as found in the 4 subtypes of nonpyramidal cells. Some of the artificial cut ends generated in the slice preparation were artificially extended using the rules of dendritic dimension shown in Fig. 2b and 4b ( Supplementary Fig. 3 ). All models were passive (Membrane resistance: 25,000 Ωcm2, intracellular resistance: 200 Ωcm, membrane capacity: 100 F/cm2)28. The equilibrium potential for the leak conductance was set to -58 mV (MA cell), -72 mV (FS cell), -62 mV (DB cell) and -63 mV (LB cell), values which reflect the experimentally determined average resting membrane potential of each of these neuron subtypes14,29. Synaptic currents were simulated as α-synapses (2 nS, τ = 1 ms) and injected in all dendritic compartments of the model cells. To model dendrites with elliptical cross sections using NEURON (which uses cylindrical cables), we first modeled each nonpyramidal neuron with tapering cylinders with cross sections matching the real morphology and then modified the leak conductance and membrane capacitance in each compartment such that they were equivalent to those estimated in our elliptical dendrites ( Fig. 3e ). The correction factors were calculated in the following steps.
1. We obtained functions from EM data by the linear regression that yield cross sectional area (S) and circumference (L) from summed length of distal dendrites (R).
DB: S = 365.83×R+122000, L = 1.5195×R+1540.9
MA: S = 1508.7×R+6958.1, L = 4.6152×R+1511.3
FS: S = 831.89×R+83164, L = 3.2663×R+1600.4
LB: S = 890.59×R+43202, L = 4.1012×R+1161.2
2. Model diameters (D) were calculated with sectional area from EM or above functions if no EM data were available.
3. To let the circular models to be equivalent to ones that have elliptical morphology, we corrected passive leak conductance and membrane capacitance by multiplying them by a correction factor L/(πD) to make the two properties larger.
We also constructed “conductance mismatch” models of each neuron by making the diameters of smaller daughter dendrites 1.5 times larger than the original size (i.e. cross-sectional area become 2.25 times larger than the original) and the diameter of larger daughter dendrites half of the original sizes (i.e. the cross-sectional area became quarter size. The conductance-matching indexes of the conductance mismatch models had larger standard deviations and wider ranges than models having realistic morphologies, although the averages were similar ( Table 2 ).
To measure the volume and surface area of somata and dendritic branches of the model cells, branches were assumed to be truncated cones in which volume (Vb) is calculated with:

Where Lb is length of the branch, ap is cross-sectional area of the proximal side, ad is cross-sectional area of distal side. Surface area (Sb) is calculated with:

Where cp is circumference of proximal side and cd is circumference of distal side. The soma was also assumed to be elliptical; prolate spheroid which volume (Vs) is calculated with:

Where a is the equatorial radius (along the x and y axes) and b is the polar radius measured under light microscope. Surface area (Ss) is calculated with:

Where  .
.
Ethics Statement
All experiments were conducted in compliance with the guidelines for animal experimentation of the Okazaki National Research Institutes.
References
Rinzel, J. & Rall, W. Transient response in a dendritic neuron model for current injected at one branch. Biophys J 14, 759–790 (1974).
Rall, W. & Rinzel, J. Branch input resistance and steady attenuation for input to one branch of a dendritic neuron model. Biophys J 13, 648–687 (1973).
Rall, W. Branching dendritic trees and motoneuron membrane resistivity. Exp Neurol 1, 491–527 (1959).
Rall, W. in Handbook of Physiology. The Nervous System. Cellular Biology of Neurons. Vol. 1 39–97 (Am. Physiol. Soc., 1977).
Jack, J. J. B., Noble, D. & Tsien, R. W. Electric Current Flow in Excitable Cells. (Oxford University Press, 1975).
Jaffe, D. B. & Carnevale, N. T. Passive normalization of synaptic integration influenced by dendritic architecture. J Neurophysiol 82, 3268–3285 (1999).
Cuntz, H., Borst, A. & Segev, I. Optimization principles of dendritic structure. Theor Biol Med Model 4, 21 (2007).
Mainen, Z. F. & Sejnowski, T. J. Influence of dendritic structure on firing pattern in model neocortical neurons. Nature 382, 363–366, 10.1038/382363a0 (1996).
Schaefer, A. T., Larkum, M. E., Sakmann, B. & Roth, A. Coincidence detection in pyramidal neurons is tuned by their dendritic branching pattern. J Neurophysiol 89, 3143–3154.
van Ooyen, A., Duijnhouwer, J., Remme, M. W. & van Pelt, J. The effect of dendritic topology on firing patterns in model neurons. Network 13, 311–325 (2002).
van Elburg, R. A. & van Ooyen, A. Impact of dendritic size and dendritic topology on burst firing in pyramidal cells. PLoS Comput Biol 6, e1000781 (2010).
Kubota, Y., Hatada, S., Kondo, S., Karube, F. & Kawaguchi, Y. Neocortical inhibitory terminals innervate dendritic spines targeted by thalamocortical afferents. J Neurosci 27, 1139–1150 (2007).
Kawaguchi, Y. & Kubota, Y. GABAergic cell subtypes and their synaptic connections in rat frontal cortex. Cereb Cortex 7, 476–486 (1997).
Kawaguchi, Y. & Kubota, Y. Neurochemical features and synaptic connections of large physiologically-identified GABAergic cells in the rat frontal cortex. Neuroscience 85, 677–701 (1998).
Kawaguchi, Y., Karube, F. & Kubota, Y. Dendritic branch typing and spine expression patterns in cortical nonpyramidal cells. Cereb Cortex 16, 696–711 (2006).
Karube, F., Kubota, Y. & Kawaguchi, Y. Axon branching and synaptic bouton phenotypes in GABAergic nonpyramidal cell subtypes. J Neurosci 24, 2853–2865 (2004).
Yamashita, A., Valkova, K., Gonchar, Y. & Burkhalter, A. Rearrangement of synaptic connections with inhibitory neurons in developing mouse visual cortex. J Comp Neurol 464, 426–437 (2003).
Behabadi, B. F. & Mel, B. W. J4 at sweet 16: a new wrinkle? Neural Comput 19, 2865–2870 (2007).
Rall, W. Electrophysiology of a dendritic neuron model. Biophys J 2, 145–167 (1962).
Rall, W. Theory of physiological properties of dendrites. Ann N Y Acad Sci 96, 1071–1092 (1962).
Houchin, J., Maxwell, D. J., Fyffe, R. E. & Brown, A. G. Light and electron microscopy of dorsal spinocerebellar tract neurones in the cat: an intracellular horseradish peroxidase study. Q J Exp Physiol 68, 719–732 (1983).
Rose, P. K., Keirstead, S. A. & Vanner, S. J. A quantitative analysis of the geometry of cat motoneurons innervating neck and shoulder muscles. J Comp Neurol 239, 89–107 (1985).
Ulfhake, B. & Kellerth, J. O. A quantitative light microscopic study of the dendrites of cat spinal alpha-motoneurons after intracellular staining with horseradish peroxidase. J Comp Neurol 202, 571–583 (1981).
Zomorrodi, R., Ferecsko, A. S., Kovacs, K., Kroger, H. & Timofeev, I. Analysis of morphological features of thalamocortical neurons from the ventroposterolateral nucleus of the cat. J Comp Neurol 518, 3541–3556 (2010).
Goldberg, J. H., Tamas, G., Aronov, D. & Yuste, R. Calcium microdomains in aspiny dendrites. Neuron 40, 807–821 (2003).
Kubota, Y., Hatada, S. N. & Kawaguchi, Y. Important factors for the three-dimensional reconstruction of neuronal structures from serial ultrathin sections. Front Neural Circuits 3, 4 (2009).
Hines, M. L. & Carnevale, N. T. The NEURON simulation environment. Neural Comput 9, 1179–1209 (1997).
Traub, R. D. et al. Single-column thalamocortical network model exhibiting gamma oscillations, sleep spindles and epileptogenic bursts. J Neurophysiol 93, 2194–2232 (2005).
Kawaguchi, Y. & Kubota, Y. Physiological and morphological identification of somatostatin- or vasoactive intestinal polypeptide-containing cells among GABAergic cell subtypes in rat frontal cortex. J Neurosci 16, 2701–2715 (1996).
Acknowledgements
We thank Y. Itoh, N. Yamaguchi, S Hatada and A. Sekigawa for technical assistance. Dr. R Benoit for generous gifts of somatostatin antiserums. Dr. Maki K. Yamada for fruitful advice to visualize microtubule. We thank Drs. Nelson Spruston, Charles J Wilson, Rafael Yuste and Jesse H Goldberg for valuable comments on the manuscript. This work was supported by KAKENHI (22120518, 20500300); Hayama Center for Advanced Studies (HCAS); and the Naito Foundation Subsidy for Natural Science Researches.
Author information
Authors and Affiliations
Contributions
Y. Kubota designed the project and conducted the electron microscopy. Y. Kawaguchi did all of slice experiments. F. Karube did LM reconstruction and data analysis. Y Kubota and F. Karube reconstruct images from serial EM photos. M. Nomura conducted all simulation studies and Y. Kubota analyzed the results. A. Mochizuki provided analysis for Fig. 4e . A.T. Gulledge aided in data analysis and interpretation. A. Schertel operated FIB/SEM for serial image data set acquisition. All authors wrote the paper.
Dendritic bifurcation conserves the cross sectional area and obeys Rall’s 3/2 power law.
(a) Reconstructed dendritic branch point in the large basket cell. Circumferences and cross-sectional areas were calculated as averages from a unit dendritic length. (b) The cross-sectional areas of parent dendrites are quite similar to the summated cross-sectional areas of daughter dendrites. Line indicates a linear fit and doted line indicates a line of unity. R: correlation coefficient to the fitted line, RU: correlation coefficient to the line of unity. (c) Drawing indicates microtubule distribution at a branching point. The microtubules found in the mother dendrite run into one of the daughter dendrites. (d) The majority of branches had near-perfect matching of parent-to-daughter conductance ( ). Line indicates linear fit and doted line indicates a line of unity. R: correlation coefficient to the fitted line, RU: correlation coefficient to the line of unity. (e) Straight lines indicate the range of possible values for two daughter dendrites having cross-sectional areas summing to match the measured value of the parent dendrite (area matching line: AM). Arcs with the same color as straight lines indicate the range of values for two daughter dendrite cross-sectional areas that follow Rall’s conductance-matching “3/2 power” law (conductance-matching curve: CM). These two lines are similar for small values typical of dendritic dimensions. The measured cross-sectional areas for each of the daughter dendrites are indicated by circles color-coded to the lines and arcs and most fall within the range of values expected for these two relationships. The minimum distance to AM and CM are shown.
). Line indicates linear fit and doted line indicates a line of unity. R: correlation coefficient to the fitted line, RU: correlation coefficient to the line of unity. (e) Straight lines indicate the range of possible values for two daughter dendrites having cross-sectional areas summing to match the measured value of the parent dendrite (area matching line: AM). Arcs with the same color as straight lines indicate the range of values for two daughter dendrite cross-sectional areas that follow Rall’s conductance-matching “3/2 power” law (conductance-matching curve: CM). These two lines are similar for small values typical of dendritic dimensions. The measured cross-sectional areas for each of the daughter dendrites are indicated by circles color-coded to the lines and arcs and most fall within the range of values expected for these two relationships. The minimum distance to AM and CM are shown.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Video 1
Supplementary Information
Supplementary Video 2
Supplementary Information
Supplementary Video 3
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Kubota, Y., Karube, F., Nomura, M. et al. Conserved properties of dendritic trees in four cortical interneuron subtypes. Sci Rep 1, 89 (2011). https://doi.org/10.1038/srep00089
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep00089
This article is cited by
-
Deep learning-driven adaptive optics for single-molecule localization microscopy
Nature Methods (2023)
-
Exact solutions to cable equations in branching neurons with tapering dendrites
The Journal of Mathematical Neuroscience (2020)
-
Neurodevelopmental Disorders: Functional Role of Ambra1 in Autism and Schizophrenia
Molecular Neurobiology (2019)
-
Spatial registration of neuron morphologies based on maximization of volume overlap
BMC Bioinformatics (2018)
-
Sculpting light to reveal brain function
Nature Neuroscience (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.