Abstract
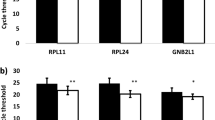
Coupling array technology to laser capture microdissection (LCM) has the potential to yield gene expression profiles of specific cell populations within tissue. However, remaining problems with linear amplification preclude accurate expression profiling when using the low nanogram amounts of RNA recovered after LCM of human tissue. We describe a novel robust method to reliably amplify RNA after LCM, allowing direct probing of 12K gene arrays. The fidelity of amplification was demonstrated by comparing the ability of amplified RNA (aRNA) versus that of native RNA to identify differentially expressed genes between two different cell lines, demonstrating a 99.3% concordance between observations. Array findings were validated by quantitative polymerase chain reaction analysis of a randomly selected subset of 32 genes. Using LCM to recover normal (N=5 subjects) or cancer (N=3) cell populations from intact human prostate tissue, three differentially expressed genes were identified. Independent investigators have previously identified differential expression of two of these three genes, hepsin and beta-microseminoprotein, in prostate cancer. Taken together, the current study demonstrates that accurate gene expression profiling can readily be performed on specific cell populations present within complex tissue. It also demonstrates that this approach efficiently identifies biologically relevant genes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 4 print issues and online access
$259.00 per year
only $64.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Schena M, Shalon D, Davis RW, Brown PO . Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995; 270: 467–470.
Schena M, Shalon D, Heller R, Chai A, Brown PO, Davis RW . Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc Natl Acad Sci USA 1996; 93: 10614–10619.
DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M et al. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet 1996; 14: 457–460.
Heller RA, Schena M, Chai A, Shalon D, Bedilion T, Gilmore J et al. Discovery and analysis of inflammatory disease-related genes using cDNA microarrays. Proc Natl Acad Sci USA 1997; 94: 2150–2155.
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA et al. Molecular portraits of human breast tumours. Nature 2000; 406: 747–752.
Welsh JB, Zarrinkar PP, Sapinoso LM, Kern SG, Behling CA, Monk BJ et al. Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proc Natl Acad Sci USA 2001; 98: 1176–1181.
Dudley AM, Aach J, Steffen MA, Church GM . Measuring absolute expression with microarrays with a calibrated reference sample and an extended signal intensity range. Proc Natl Acad Sci USA 2002; 99: 7554–7559.
Yue H, Eastman PS, Wang BB, Minor J, Doctolero MH, Nuttall RL et al. An evaluation of the performance of cDNA microarrays for detecting changes in global mRNA expression. Nucleic Acids Res 2001; 29: E41–1.
Iscove NN, Barbara M, Gu M, Gibson M, Modi C, Winegarden N . Representation is faithfully preserved in global cDNA amplified exponentially from sub-picogram quantities of mRNA. Nat Biotechnol 2002; 20: 940–943.
Puskas LG, Zvara A, Hackler Jr L, Van Hummelen P . RNA amplification results in reproducible microarray data with slight ratio bias. Biotechniques 2002; 32: 1330–1334, 1336, 1338, 1340.
Smith L, Underhill P, Pritchard C, Tymowska-Lalanne Z, Abdul-Hussein S, Hilton H et al. Single primer amplification (SPA) of cDNA for microarray expression analysis. Nucleic Acids Res 2003; 31: e9.
Pabon C, Modrusan Z, Ruvolo MV, Coleman IM, Daniel S, Yue H et al. Optimized T7 amplification system for microarray analysis. Biotechniques 2001; 31: 874–879.
Wang E, Miller LD, Ohnmacht GA, Liu ET, Marincola FM . High-fidelity mRNA amplification for gene profiling. Nat Biotechnol 2000; 18: 457–459.
Wadenback J, Clapham DH, Craig D, Sederoff R, Peter GF, von Arnold S et al. Comparison of standard exponential and linear techniques to amplify small cDNA samples for microarrays. BMC Genom 2005; 6: 61.
Huang X, Chen S, Xu L, Liu Y, Deb DK, Platanias LC et al. Genistein inhibits p38 map kinase activation, matrix metalloproteinase type 2, and cell invasion in human prostate epithelial cells. Cancer Res 2005; 65: 3470–3478.
Hayes SA, Huang X, Kambhampati S, Platanias LC, Bergan RC . p38 MAP kinase modulates Smad-dependent changes in human prostate cell adhesion. Oncogene 2003; 22: 4841–4850.
Liu Y, Jovanovic B, Pins M, Lee C, Bergan RC . Over expression of endoglin in human prostate cancer suppresses cell detachment, migration and invasion. Oncogene 2002; 21: 8272–8281.
Schwartz GN, Liu YQ, Tisdale J, Walshe K, Fowler D, Gress R et al. Growth inhibition of chronic myelogenous leukemia cells by ODN-1, an aptameric inhibitor of p210bcr-abl tyrosine kinase activity. Antisense Nucleic Acid Drug Dev 1998; 8: 329–339.
Lin Y NS, Lan H, Attie AD, Yandell BS . A daptive gene picking with microarray data: detecting important low abundance signals. In: G Parmigiani EG, RA Irizarry, SL Zeger (eds). The Analysis of Gene Expression Data: Methods and Software. Springer-Verlag: New York, 2003.
Kyle E, Neckers L, Takimoto C, Curt G, Bergan R . Genistein-induced apoptosis of prostate cancer cells is preceded by a specific decrease in focal adhesion kinase activity. Mol Pharmacol 1997; 51: 193–200.
Rohlff C, Blagosklonny MV, Kyle E, Kesari A, Kim IY, Zelner DJ et al. Prostate cancer cell growth inhibition by tamoxifen is associated with inhibition of protein kinase C and induction of p21(waf1/cip1). Prostate 1998; 37: 51–59.
Jovanovic BD, Huang S, Liu Y, Naguib KN, Bergan RC . A simple analysis of gene expression and variability in gene arrays based on repeated observations. Am J Pharmacogenom 2001; 1: 145–152.
Baugh LR, Hill AA, Brown EL, Hunter CP . Quantitative analysis of mRNA amplification by in vitro transcription. Nucleic Acids Res 2001; 29: E29.
Goley EM, Anderson SJ, Menard C, Chuang E, Lu X, Tofilon PJ et al. Microarray analysis in clinical oncology: pre-clinical optimization using needle core biopsies from xenograft tumors. BMC Cancer 2004; 4: 20.
Gomes LI, Silva RL, Stolf BS, Cristo EB, Hirata R, Soares FA et al. Comparative analysis of amplified and nonamplified RNA for hybridization in cDNA microarray. Anal Biochem 2003; 321: 244–251.
Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR et al. Laser capture microdissection [see comments]. Science 1996; 274: 998–1001.
Shah RB, Mehra R, Chinnaiyan AM, Shen R, Ghosh D, Zhou M et al. Androgen-independent prostate cancer is a heterogeneous group of diseases: lessons from a rapid autopsy program. Cancer Res 2004; 64: 9209–9216.
Garde SV, Basrur VS, Li L, Finkelman MA, Krishan A, Wellham L et al. Prostate secretory protein (PSP94) suppresses the growth of androgen-independent prostate cancer cell line (PC3) and xenografts by inducing apoptosis. Prostate 1999; 38: 118–125.
Tsurusaki T, Koji T, Sakai H, Kanetake H, Nakane PK, Saito Y . Cellular expression of beta-microseminoprotein (beta-MSP) mRNA and its protein in untreated prostate cancer. Prostate 1998; 35: 109–116.
Chan PS, Chan LW, Xuan JW, Chin JL, Choi HL, Chan FL . In situ hybridization study of PSP94 (prostatic secretory protein of 94 amino acids) expression in human prostates. Prostate 1999; 41: 99–109.
Klezovitch O, Chevillet J, Mirosevich J, Roberts RL, Matusik RJ, Vasioukhin V . Hepsin promotes prostate cancer progression and metastasis. Cancer Cell 2004; 6: 185–195.
Magee JA, Araki T, Patil S, Ehrig T, True L, Humphrey PA et al. Expression profiling reveals hepsin overexpression in prostate cancer. Cancer Res 2001; 61: 5692–5696.
Kenzelmann M, Klaren R, Hergenhahn M, Bonrouhi M, Grone HJ, Schmid W et al. High-accuracy amplification of nanogram total RNA amounts for gene profiling. Genomics 2004; 83: 550–558.
Mutsuga N, Shahar T, Verbalis JG, Xiang CC, Brownstein MJ, Gainer H . Regulation of gene expression in magnocellular neurons in rat supraoptic nucleus during sustained hypoosmolality. Endocrinology 2005; 143: 1254–1267.
Alevizos I, Mahadevappa M, Zhang X, Ohyama H, Kohno Y, Posner M et al. Oral cancer in vivo gene expression profiling assisted by laser capture microdissection and microarray analysis. Oncogene 2001; 20: 6196–6204.
Jenson SD, Robetorye RS, Bohling SD, Schumacher JA, Morgan JW, Lim MS et al. Validation of cDNA microarray gene expression data obtained from linearly amplified RNA. Mol Pathol 2003; 56: 307–312.
Luzzi V, Mahadevappa M, Raja R, Warrington JA, Watson MA . Accurate and reproducible gene expression profiles from laser capture microdissection, transcript amplification, and high density oligonucleotide microarray analysis. J Mol Diagn 2003; 5: 9–14.
Wu MS, Lin YS, Chang YT, Shun CT, Lin MT, Lin JT . Gene expression profiling of gastric cancer by microarray combined with laser capture microdissection. World J Gastroenterol 2005; 11: 7405–7412.
Crnogorac-Jurcevic T, Efthimiou E, Nielsen T, Loader J, Terris B, Stamp G et al. Expression profiling of microdissected pancreatic adenocarcinomas. Oncogene 2002; 21: 4587–4594.
Leethanakul C, Patel V, Gillespie J, Shillitoe E, Kellman RM, Ensley JF et al. Gene expression profiles in squamous cell carcinomas of the oral cavity: use of laser capture microdissection for the construction and analysis of stage-specific cDNA libraries. Oral Oncol 2000; 36: 474–483.
Febbo PG, Thorner A, Rubin MA, Loda M, Kantoff PW, Oh WK et al. Application of oligonucleotide microarrays to assess the biological effects of neoadjuvant imatinib mesylate treatment for localized prostate cancer. Clin Cancer Res 2006; 12: 152–158.
Stoyanova R, Upson JJ, Patriotis C, Ross EA, Henske EP, Datta K et al. Use of RNA amplification in the optimal characterization of global gene expression using cDNA microarrays. J Cell Physiol 2004; 201: 359–365.
Mutsuga N, Shahar T, Verbalis JG, Xiang CC, Brownstein MJ, Gainer H . Regulation of gene expression in magnocellular neurons in rat supraoptic nucleus during sustained hypoosmolality. Endocrinology 2005; 146: 1254–1267.
Arbeitman MN, Furlong EE, Imam F, Johnson E, Null BH, Baker BS et al. Gene expression during the life cycle of Drosophila melanogaster. Science 2002; 297: 2270–2275.
Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV et al. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol 2003; 20: 1377–1419.
Kelley MJ, Glaser EM, Herdon JE, Becker F, Santella RM, Hecht SS et al. Safety and efficacy of weekly oral oltipraz in chronic smokers. Cancer Epidemiol Biomarkers Prev 2005; 14: 892–899.
Zhao H, Hastie T, Whitfield ML, Borresen-Dale AL, Jeffrey SS . Optimization and evaluation of T7 based RNA linear amplification protocols for cDNA microarray analysis. BMC Genom 2002; 3: 31.
Schneider J, Buness A, Huber W, Volz J, Kioschis P, Hafner M et al. Systematic analysis of T7 RNA polymerase based in vitro linear RNA amplification for use in microarray experiments. BMC Genom 2004; 5: 29.
Jovanovic BD, Helenowski IB, Rademaker AW, Bedford DF, Bergan RB . Some statistical issues in study of variability of gene expression with applications in prostate cancer study. J Probab Statist Quant Manage 2004; 1: 51–60.
Jovanovic BD, Bergan RC, Kibbe WA . Some aspects of analysis of gene array data. Cancer Treat Res 2002; 113: 71–89.
Acknowledgements
This work was supported by the National Cancer Institute (CA099263, CA37403, and Specialized Program of Research Excellence grant CA90386, to RCB), and a National Center for Research Resources Grant (M01 RR00048), both from the National Institutes of Health, Department of Health and Human Services
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ding, Y., Xu, L., Chen, S. et al. Characterization of a method for profiling gene expression in cells recovered from intact human prostate tissue using RNA linear amplification. Prostate Cancer Prostatic Dis 9, 379–391 (2006). https://doi.org/10.1038/sj.pcan.4500888
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.pcan.4500888
Keywords
This article is cited by
-
Identification of markers of prostate cancer progression using candidate gene expression
British Journal of Cancer (2012)
-
Endoglin suppresses human prostate cancer metastasis
Clinical & Experimental Metastasis (2011)
-
Evaluation of methods for amplification of picogram amounts of total RNA for whole genome expression profiling
BMC Genomics (2009)
-
Endoglin inhibits prostate cancer motility via activation of the ALK2-Smad1 pathway
Oncogene (2007)