Abstract
Patterns of pollen dispersal are central to both the ecology and evolution of plant populations. However, the mechanisms controlling either the dispersal process itself or our estimation of that process may be influenced by site-specific factors such as local forest structure and nonuniform adult genetic structure. Here, we present an extension of the AMOVA model applied to the recently developed TWOGENER analysis of pollen pool structure. This model, dubbed the Stepwise AMOVA (StAMOVA), focuses on determining to what extent ecological, demographic, and/or environmental factors influence the observed genetic variation in spatially separated pollen pools. The analysis is verified for efficacy, using an extensive battery of simulations, illustrating: (1) how nonuniform adult genetic structure influences the differentiation of spatially separated pollen pools, and (2) how effectively the Stepwise analysis performs in carrying out the appropriate corrections. Finally, the model is applied to a Quercus alba data set, from which we have prior evidence that the adult genetic structure is nonuniformly distributed across the sampling landscape. From this data set, we show how the Stepwise model can be applied to remove the effects of spatial adult genetic structure on pollen pool differentiation and contrast these results with those derived from the original TWOGENER analysis.
Similar content being viewed by others
Introduction
Pollen-mediated gene movement plays a critical role in the evolutionary dynamics of plant populations (Harper, 1977). To understand the process of pollen-mediated gene movement, we must understand the extent to which adult spatial, ecological, and genetic heterogeneity might influence the mechanisms of pollen dispersal as well as our estimations of pollen dispersal distances. Theoretical models describing wind-dispersed pollen typically assume a highly skewed, or leptokurtic, form (eg, Levin and Kerster, 1974; Okubo and Levin, 1989). Smouse et al (2001) recently proposed the TWOGENER analysis that utilizes spatially separated mothers to sample the pollen pool, with the aim of quantifying ongoing gene flow in terms of the mean pollen dispersal distance and the genetic effective number of pollen donors. Under this model, any increase in dispersal distance will result in lower among-mother genetic divergence in sampled pollen pools.
Assuming specific pollen dispersal distributions, Austerlitz and Smouse (2001a) show that the mean pollen dispersal distance (δ) can be derived from the TWOGENER analysis and is inversely proportional to the observed genetic structure of spatially separated pollen pools (ΦFT). Furthermore, Austerlitz and Smouse (2001b) showed that while fine-scale adult structure, as is often found in plant populations, can influence our estimates of dispersal distance, the overall magnitude of spatially autocorrelated adults is rather small. All of these models estimate pollen movement as though it were similar across all individuals, a supposition that is unlikely to be true in natural populations. In fact, one might expect extensive heterogeneity of the dispersal environment, due both to ecological and genetic factors that vary across the landscape. However, we do not know whether such heterogeneity actually affects the process of pollen movement, and if so, what is the extent that effect has on our estimates.
One factor that can influence genetic differentiation among spatially separated pollen pools is heterogeneity in the distance that the pollen is dispersed. Both intrinsic factors, such as pollen morphology, release height, and settling velocity (Levin and Kerster, 1974), as well as external factors such as wind direction and speed, local density of pollen donors, and forest structure (Dyer and Sork, 2001) can significantly influence pollen dispersal distance. The external factors have a greater opportunity to be nonuniformly distributed across the landscape, causing a greater influence on pollen dispersal distance. For example, Okubo and Levin, 1989, provide a theoretical framework within which dispersal distance can be characterized in terms of diffusion processes and advection, taking into consideration the effects of local air turbulence. Their models predict that increases in local air turbulence will result in a significant reduction in pollen dispersal distance. Consistent with these hypotheses, Dyer and Sork, (2002) show that genetic diversity within spatially separated pollen pools for Pinus echinata is inversely related to the density of all canopy species and independent of P. echinata density. They argue that the high density of all canopy species on site impedes the movement of pollen both within as well as into the stand. If true, then dispersal distance is not constant across the landscape and may result in an increase in the among-mother component of variance because the mothers are sampling different-sized pollen donor populations.
The distribution and abundance of local pollen donors can also influence the genetic structure of pollen pools. Austerlitz and Smouse (2001a) have shown that ΦFT is insensitive to changes in the spatial separation of sampled mothers, provided that the separation of mothers is sufficiently large, ∼5δ, where δ is the average distance that the pollen is dispersed. However, clumping or nonuniform distributions of pollen donors can have a significant influence on spatial genetic structure (eg, Dolgez et al, 1998). Further, several studies of forest tree mating systems have reported the effects of local pollen donor density on pollen pool composition (eg, Farris and Mitton, 1984; Knowles et al, 1987; Shea, 1987; Murawski and Hamrick, 1991). Additional factors such as flowering phenology (Sampson et al, 1990), adult allele frequency gradients (Dyer and Sork, 2002), elevation gradients (Loechelt and Franke, 1996), and inbreeding and spatial autocorrelation among adult individuals (Austerlitz and Smouse, 2001b) also act to increase the genetic divergence among sampled pollen pools. If any of these factors have a significant influence on pollen pool composition, then estimates of pollen dispersal distance will be biased downward. Our understanding of the role of pollen-mediated gene movement in the evolutionary dynamics of plant populations will be sharpened by identifying those factors that influence pollen dispersal, either directly through modification of dispersal distance or indirectly as a result of local demographic or genetic processes.
In this paper, we present a multivariate analysis of pollen pool differentiation based on the TWOGENER analysis. This novel model, dubbed the Stepwise Analysis of MOlecular VAriance (StAMOVA) identifies and partitions out the effects of external variables on pollen pool differentiation. In doing so, this model provides a robust statistical methodology, by means of a general linear model, which quantifies the effects that ecological and spatial covariates have on the composition of spatially separated pollen pools. Following an introduction to the analysis, we evaluate its effectiveness in detecting one category of external factors that can significantly influence the pollen pool genetic structure, the nonuniform distribution of adult genetic structure. The effects of broad-scale adult genetic structure on pollen pool composition were examined via an extensive battery of simulations, wherein a multilocus genetic structure is imposed upon the pollen donor population. We then contrast the results of the Stepwise analysis with those provided by the TWOGENER analysis. Finally, this analysis is applied to the Quercus alba data set presented in the original TWOGENER paper (Smouse et al, 2001), where previous analyses have identified multivariate genetic structures in the adult population. We show that removing the effects imposed on the observed differentiation in sampled pollen pools by the gradient in the adult genetic structure not only increases the accuracy of dispersal distance estimates but also serves as a signal to the investigator of an underlying nonuniformity in the adult genetic structure. We close by commenting on the general applicability of the StAMOVA model for the analysis of the population genetic structure. While this model is currently being applied to contemporary pollen movement dynamics under the TWOGENER model, the StAMOVA is just as readily applied to analyses of adult genetic structure, a topic we return to in a later manuscript (Dyer et al, in preparation).
Methods
The main goal here is to use the observed genetic structure of spatially separated pollen pools to draw inferences about the mean pollen dispersal distances and effective pollination neighborhood sizes. However, the observed differentiation of spatially separated pollen pools may be a function of factors other than the distance that pollen is dispersed from the pollen donor. For example, if the pollen donor population has a gradient in allele frequencies (across elevation for example), the differentiation of pollen pools will be exacerbated, because maternal trees separated along the gradient will be drawing from pollen donor pools that differ systematically in their allele frequency profiles (Dyer and Sork, 2002). The task at hand is to develop a method that allows us to determine the extent to which the observed genetic variation among sampled pollen pools is due to pollen dispersal distance, as opposed to factors either affecting the dispersal distance directly, as is expected for spatial variation in stand density, or indirectly, as would be expected from changes resulting from an allele frequency gradient.
The Stepwise analysis is a multivariate general linear model conforming to the AMOVA analysis of Excoffier et al (1992). In fact, the AMOVA analysis is a special case of the more general Stepwise model, where one is only interested in decomposing the genetic variance into within- and among-strata components. For this reason, we abbreviate the Stepwise model as StAMOVA to draw attention to its extension of the original AMOVA model. The TWOGENER analysis of pollen pool structure (Smouse et al, 2001) also utilizes the AMOVA model for the analysis of pollen structure.
In general terms, the AMOVA analysis (and the TWOGENER analysis as well as the StAMOVA by extension) relies upon the multilocus pair-wise genetic distance matrix, D. The D matrix has the form:

where δij2 is the additive, pair-wise genetic distance between the ith and jth pollen haplotypes. The elements of the D matrix, δij2, are 0 when i=j, and may be non-negative when i≠j, the exact value of which depends upon the pollen haplotypes i and j. The StAMOVA model does not use distance matrices, rather it focuses on variance/covariance matrices in a more traditional statistical sense. The D matrix can easily be transformed into a genetic variance/covariance matrix, following Gower (1966). We refer to this variance–covariance matrix as YY′, and specifically use this notation to draw attention to the fact that it is identical to the Sums of Squares and CrossProducts (SSCP) matrix in a Q-mode analysis (ie, the differences between N individuals, not p-variables). The sums of squares and crossproducts for traditional multivariate analysis are estimated from the Y′Y matrix (SSCP is p × p in size; an R-mode analysis), whereas we have the YY′ matrix (SSCP is N × N; Q-mode analysis). The overall variation in either SSCP matrix is given by the trace, tr[]. Since tr[XY]=tr[YX], the analysis can be carried out using the outer-product matrix (YY′) in exactly the same way as the traditional multivariate analysis does with the inner-product matrix (Y′Y). It should be noted that much larger matrices are being manipulated in Q-mode, requiring more computational resources for large data sets. The code for this analysis is provided and is available from RJD upon request.
The predicted values of any linear model are given by

where the (N × N) matrix H is symmetrical and idempotent, given by

(Johnson and Wichern, 1992), with X being the multivariate design matrix. The structure of X follows that for a traditional analysis of dummy variables among K strata (see Johnson and Wichern, 1992).
Returning to decomposition of the linear model, the outer product matrix of predicted values, ŶŶ′, can now be estimated as

following [1]. Since the H matrix is idempotent (ie, HH=H′=(H)−1=H), we can drop the second multiplication for the purposes of computational efficiency. The outer product matrix of the residuals (ie, that variance/covariance matrix not accounted for by the model up to this point) is

where I is the N × N identity matrix.
Using the trace of the outer-product matrices, we decompose the total variation among pollen haplotypes into components representing the variation explained by the model and the remaining variation (ie, the residual or error variance) as:

The outer-product matrix formulation outlined above translates the AMOVA framework used in the TWOGENER analysis directly into its underlying multivariate linear model form. With this formulation, we not only retain the ability to extract the exact same total, model, and residual sums of squares from the analysis but we also gain the advantages of a generalized linear model, from which we may extract additional information in terms of covariates as well as construct sampling designs more detailed than simple hierarchical nesting of populations within regions.
Stepwise decomposition of variation
Adopting a stepwise approach (Draper and Smith, 1981; Searle, 1997) allows assessment of the variation in the observed pollen pools that can be accounted for by external factors, such as maternal position on the landscape (assessing the effects of allele frequency gradients in the adult population) or ecological variables (such as stand characteristics). Following the removal of variation explained by external variables, we then estimate the among- and within-strata components yielding a Φ-statistic corrected for the influence of external variables. This stepwise treatment follows Henderson's Method 2 procedure, as outlined in Searle (1997).
To demonstrate the stepwise approach, consider the case where only one external predictor variable is influencing pollen pool composition. For the purposes of this discussion, consider the maternal y-coordinate, as the simulations conducted below impose an allele frequency gradient on the adults, along the y-axis. First, standardize the y-coordinate to mean zero to remove the necessity of an intercept term. Call this vector XE where the subscript E denotes the external, predictor variables, assumed to be fixed effects and measured without error. Substituting XE into (1) provides the external hypothesis matrix, HE. Substituting HE into (2) and (3), the trace of the component matrices of (5) estimates the variation explained by the predictor variables, tr[ŶŶ′], as well as the residual variation not accounted for by the predictor variables. The significance of the XE is evaluated by examining the reduction in the sums of squares, denoted R(XE), attributable to that variable. The reduction in the sums of squares due to fitting XE to the model is:

Under the null hypothesis of no variation in the pollen pool, other than that attributable to pollen dispersal distance, the expectation is that the residual variation is zero, E[R(XE)]=0. In other words, the null hypothesis states that the reduction in the sums of squares associated with XE is invariant with respect to the pairing of pollen donor haplotypes and the elements of XE. If the null hypothesis were true, the observed value of R(XE) would be equally likely under random permutation of XE among strata. Therefore, the null distribution of R(XE) is constructed by permuting the elements of XE among sampled females and recalculating the R(XE-Perm). Comparing the observed R(XE) to the distribution of permuted R(XE-Perm) allows determination of a ‘tail probability’ significance to be ascertained. This is exactly the same procedure for testing the significance of population level differences in the original AMOVA analysis. If there is more than one external variable of interest (ie, if XE is (N × G) with G>1), this framework is easily extended by simply increasing the number of predictor variable columns in the XE matrix. Higher-order polynomials and interaction terms are added just as in a regular multiple regression analysis (Draper and Smith, 1981).
Once the relationships between the external variables and the pollen donor haplotypes have been established, the maternal design matrix, XM (from [3]) is entered into the model and the TWOGENER analysis is performed, following [5], with the exception that the YY′ matrix is replaced by the RERE′ matrix of residual variation, after removing the effects of external predictor variables. The reduction in the sums of squares due to fitting the mothers, following the removal of variation due to the external variables is:

Note that there is no interaction term in the model, since the sums of squares associated with the mothers (XM) is calculated from residual variation after removing the G external variable(s). Zelen (1968) and Searle (1968) equate this method to Henderson's (1953) ‘fitting constants’ formulation.
Decomposition of the variance components and estimation of the Φ-statistics from the among-mother component of variation follow the methods outlined in Excoffier et al (1992), with one exception. The error degrees of freedom need to be adjusted to take into account the external variables (G of them) that have already been entered into the model (see Table 1). If the external variables are random effects, then the variance components for these effects can be partitioned following Searle (1968).
Simulation methods to evaluate stepwise model
Simulations were used to highlight the effects that spatially organized pollen donor genotypes have on the among-mother component of genetic variation, σA2, as well as to evaluate the utility of the Stepwise model. A total of 10 000 adults were simulated on a 100 × 100 square lattice (adult density is 1 individual per map unit), following Smouse et al (2001). Pollen dispersal was simulated by drawing pollen donors from a bivariate exponential distribution with a mean pollen dispersal distance equal to 5 map units (see Smouse et al, 2001). The allele frequency gradients, ∂p, were set to p=0.00 (no gradient) and ∂p=[0.10, 0.20, 0.30] (hereafter gradient populations). In populations where an allele frequency gradient was imposed upon the adult population, the strength of the allele frequency gradient was quantified as the allele frequency change from one end of the lattice to the other. All starting allele frequencies were set to p=q=0.5, prior to the application of the gradient. All adults were assigned 10 polymorphic loci, with two alleles per locus, the frequency of which was determined by (1) the individuals spatial location along the y-axis, and (2) the strength of the allele frequency gradient, ∂p.
A total of 49 maternal trees were spread uniformly across the landscape, in seven rows of seven individuals. Each maternal individual produced 10 outcross offspring. To provide confidence intervals around the variation due to the gradient, as well as the among-mother component of genetic variation, 1000 iterations of 490 simulated matings were run and analyzed for each level of the allele frequency gradient. During each iteration, the entire adult population was recreated to randomize the placement of pollen donors.
The pollen donor haplotypes sampled by each of the maternal individuals during each simulation were analyzed using two methods. First, the TWOGENER analysis was performed under the alternative hypothesis that the differentiation among mothers was simply due to the pollen dispersal distance. Second, the Stepwise treatment was applied to the same data set, using the maternal location along the simulated allele frequency gradient as the external variable. Variance partitioning due to the spatial variable and the among-mother components were estimated as above.
Case study: Q. alba in a Missouri Ozark Forest
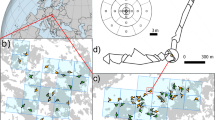
We applied the Stepwise analysis to a data set consisting of 35 maternal Q. alba trees and 1024 multilocus offspring. These data are a subset of offspring analyzed for the first TWOGENER Analysis paper (see Smouse et al, 2001). Offspring have been assayed for eight allozyme loci, yielding an exclusion probability of PE=0.707. Single-locus adult genetic structure within this population shows no significant differentiation (Koop, 1996), but multivariate analysis of adult genetic structure revealed a significant gradient in allele frequencies running roughly north–south (Figure 1; AL Koop and VL Sork, unpublished). Furthermore, Gram and Sork (1999, 2001) reported significant correlations between multilocus genetic variables and both multivariate forest structure and local population densities. The Stepwise analysis was applied to this data set, considering only the north–south gradient in allele frequencies. We first present the differentiation in sampled pollen pools in the uncorrected data set, using the TWOGENER analysis. We then present the Stepwise analysis, using the maternal north–south coordinate, hereafter ‘northing’, as the external predictor variable. We also test the east–west coordinate as a second spatial variable, although there was no a priori reason to suspect a gradient in adult genetic structure along this axis. All analyses were tested for significance by permuting offspring among mothers 1000 times and constructing the null distribution of the sampling statistic described above.
Results
The simulations show that the estimate of the among-mother component of genetic variation, σA2, increases significantly with gradients in pollen allele frequencies (Figure 2, filled boxes), as shown previously in Dyer and Sork (2001). The simulation results show a 2.5-fold increase in σA2, observed between simulated matings in populations with uniform allele frequencies and those with an allele frequency gradient of ∂p=0.30. While the overall range of allele frequency gradients, ∂p=0.00 vs 0.30, is exceedingly large and perhaps exaggerated, relative to what may be observed in natural populations, it clearly shows how the genetic constitution of the pollen donors influences the observed pollen pool differentiation.
Estimated among-mother components of variation, A2 (ie, that attributable to the effects of dispersal distance), across a nonuniform adult genetic structure gradient, ∂p=[0.00, 0.10, 0.20, 0.30]. TWOGENER results are represented as closed boxes, whereas the Stepwise results are represented by open circles.
While the bias in σA2, due to the spatial genetic heterogeneity of the adults is significant, the magnitude of the differences is relatively small when translated into standardized variance among strata, ΦFT. Translating the σA2 values in Figure 2 into ΦFT yields an absolute difference of 0.02 (two-sample t; t=38.81; P<0.001). Therefore, in the most extreme case given by these simulations, where the allele frequency change is ∂p=0.30, the calculated differentiation between pollen pools may be small. However, even with the smallest allele frequency gradient, ∂p=0.10, the increase in σA2 is large enough to differentiate ΦFT,∂p=0.00 and ΦFT,∂p=0.10 (two-samples t, t=3.402, P=0.0003). The differences in ΦFT before and after removal of the environmental ‘covariate’ are relatively small and may be of dubious biological relevance in terms of how it influences future genetic structure. However, their significance in explaining the observed distribution of pollen donor structure across the landscape is arguably more important than the magnitude of their effects. The significant retention in the StAMOVA model suggests a functional relationship that has the ability to modify the patterns and influences of pollen-mediated gene flow.
The StAMOVA analysis successfully partitioned total variation into that corresponding to the gradient in adult allele frequencies, and that due to differences among mothers (Figure 2). Without removing the effects of the allele frequency gradient in the adult population, the additional sums of squares associated with the gradient remained within the among-mother component. However, under the StAMOVA, the sums of squares are partitioned between that due to the gradient and that due to the among-mother component. As a result, the among-mother variance is unaffected by the presence of the allele frequency gradient in the adults, provided one accounts for the latter. The analysis of the external variable in simulations without an allele frequency gradient (∂p=0.00) was nonsignificant (ie, the reduction in the sums of squares due to the external variable in the cases where there was no allele frequency gradient was not significant). In these cases, it is not appropriate to retain the external variable in the model, and the StAMOVA analysis reduces to the original TWOGENER analysis. The addition of the spatial location perpendicular to the allele frequency gradient was also not significant, as would be expected since there is no systematic change in allele frequencies along this axis. With the StAMOVA, simulations where ∂p=[0.10, 0.20, 0.30] showed no significant increase in σA2 over that for ∂p=0.00, once the effects of the external variable were removed. Subsequent estimates of γ (the mean dispersal distance) were unaffected by ∂p once the spatial covariate was removed and were not significantly different than the values entered into the simulation study (data not shown).
Stepwise model applied to Q. alba
The StAMOVA was applied to a subset of the Q. alba progeny arrays used in the original TWOGENER paper (Smouse et al, 2001). Differentiation between the 35 sampled pollen pools was comparable to that from the analysis of the full data set, ΦFT=0.063 (P<0.001; Table 2a) here, vs ΦFT=0.061 in Smouse et al (2001). However, after removing the effects of maternal position along the north–south gradient, the differentiation among pollen pools was reduced to ΦFT=0.056 (P<0.001; Table 2b). The reduction in the sums of squares due to the variable northing was significant (P<0.012), but the addition of the east–west coordinate was, as expected, not significant (P>0.21; data not shown). The reduction in the differentiation among Q. alba mothers translates into a 13% increase in the estimate of the area from which each mother sampled pollen. Prior to removing the influence of the maternal north–south position in the landscape, each mother was estimated to sample pollen from donors within an area of roughly 839 m2. Following the removal of maternal position within the landscape, mapping the multivariate gradient in the adult genetic structure, each mother was estimated to sample from pollen donors within an area of 952 m2. Furthermore, removing the effects caused by northing resulted in a 6% increase in the estimate of the average pollen dispersal distance (17.4 vs 16.3 m; after Smouse et al, 2001).
Discussion
This paper introduces a novel modification of the AMOVA model applied to the TWOGENER analysis of gene flow. This model, while not limited in its application to TWOGENER-type analyses, allows the identification of, and gauges the extent to which, external variables can influence sampled genetic structure. The Stepwise model casts the AMOVA model in terms of a generalized multivariate linear model, allowing addition of any number of variables hypothesized to influence the pollen pool structure. The main benefit of adopting a linear model for the analysis of pollen pool differentiation, as we have done here for the TWOGENER analysis, is the wide range of sampling designs available to quantify the extent to which external factors can influence the distribution of genetic structure. This paper demonstrates the effects of removing the covariation between adult genetic structure and pollen pool genetic structure, but alternate designs focusing on other biological or spatial factors may just as easily be applied.
The Stepwise analysis extends the utility of the TWOGENER analysis, but retains the same underlying assumptions. The coding of multilocus genotypes for the Stepwise analysis for either offspring sampled from angiosperms, where the paternal contribution may be ambiguous when offspring and mothers share the same heterozygotic state, or for conifers, where the maternal haplotype is available within the megagametophyte, is exactly the same as presented in Smouse et al (2001; Table 1). Furthermore, the effects of genetic marker resolution, inter-female sampling distance, fine-scale adult autocorrelative structure, adult inbreeding, and pollen donor density, as presented in Smouse et al (2001) and Austerlitz and Smouse (2001a, 2001bapply to the Stepwise treatment as well.
The simulation results show that deviations from genetic uniformity of adults across the landscape can significantly bias the estimation of pollen pool differentiation. While linear allele frequency gradients were used as an example here, mostly for simplification of the simulations, any deviations from uniform adult genetic structure would yield similar results. For example, Austerlitz and Smouse (2001b) have recently shown that adult inbreeding and spatially correlated coancestry both increase pollen pool differentiation. In general terms, any factor that influences adult genetic structure in a nonuniform manner across the landscape, such as microsite selection, variation in population size, local population density, and fragmentation (Ledig, 1992; Allard et al, 1993; Ellstrand and Elam, 1993; Gram and Sork, 1999, 2001) will increase the variation among spatially separated pollen pools.
In addition to adult genetic structure, pollen pool differentiation may be influenced by a number of other factors, such as the degree of spatial heterogeneity in natural populations. For example, many authors have suggested that the density of local pollen donors can significantly influence outcrossing rates in forest trees (eg, Farris and Mitton, 1984; Knowles et al, 1987; Shea, 1987; Murawski and Hamrick, 1991). In stands with few pollen donors, the pollen pool surrounding each maternal individual will have a higher proportion of her own pollen, possibly increasing the probability of producing selfed offspring. With only a modest increase in sampling effort, both ecological and spatial variables can easily be collected for each maternal individual, and can be added to the analysis.
By incorporating these additional factors into the sampling design, the array of testable hypotheses is broadened. Instead of asking what the mean dispersal distance is for a given taxa, hypotheses targeting the factors that influence the average dispersal distance can be tested. Hypotheses of this type are becoming increasingly important for the development of conservation and management practices. For example, in natural stands of P. echinata, the density of all canopy tree species has a significant effect on pollen pool diversity, whereas the density of P. echinata individuals within the stand had no detectable effect (see Figure 4 in Dyer and Sork, 2001). A StAMOVA decomposition of the P. echinata data shows that the density of heterospecifics, in addition to being influential on the diversity of sampled pollen pools, also influences the genetic differentiation among pollen pools. Dyer and Sork (2001) originally found ΦST=0.10 for the among-site component of genetic differentiation. After removing the effects variation due to heterogeneity in physical stand architecture, ΦST was reduced to 0.09 (P=0.027, RJ Dyer, unpublished StAMOVA results). Again, the extent to which forest architecture influences pollen dispersal is small yet significant. For a species whose pollen dispersal distance is much greater than that for its seeds, such as P. echinata, these results suggest that forest structure can have significant influences on the transmission of genes. From these results, specific hypotheses can be formulated regarding how alterations of forest structure influence pollen movement, in terms of pollen pool heterogeneity, the diversity of sampled pollen donors, and the propensity to produce inbred offspring (RJ Dyer, in preparation).
The StAMOVA approach can be used to estimate the magnitude and relative importance of several factors simultaneously. The simulation and Q. alba examples presented here only highlight a single external variable of interest. However, since the StAMOVA is a general linear model, any number of additional external factors and interactions among external factors can be easily added to the model just as in a multiple regression analysis. For example, in addition to the density of local pollen donors, the degree of phenological overlap has also been shown to influence pollen pool composition significantly (eg, Sampson et al, 1990; Adams and Birkes, 1991). By sampling across a range of densities in the degrees of phenological overlap, one could isolate the importance of each of these factors as well as their interaction in shaping the distribution of genetic structure within spatially separated pollen pools.
Perhaps the most important benefit of the StAMOVA approach is highlighted by the results of the Q. alba analysis. It is clear from the partitioning of genetic variation in Table 2 that the maternal position within the landscape is predictive of differentiation among sampled pollen pools. This result is important on two counts. First, this immediately suggests that there may be an underlying adult structure. While the specific nature of the adult structure requires further inquiry, the fact remains that inferences can be drawn with respect to adult genetic structure by examination of sampled pollen pools. Second, the StAMOVA analysis allows the simultaneous analysis of the interaction between genetic and nongenetic variables. In this case, the nongenetic variable was the maternal position within the landscape. The significant covariate highlighted the extent to which the environment in which gene flow occurred influences the distribution of genetic structure. As long as the spatial separation of maternal individuals from which samples are drawn is sufficiently large, relative to the mean dispersal distance (see Austerlitz and Smouse, 2001a, 2001b), the structure in local pollen pools will reflect that of local pollen donors.
While the model presented here builds upon the original AMOVA analysis of Excoffier et al (1992), the TWOGENER formulation presented by Smouse et al (2001), as well as the theoretical framework provided by Austerlitz and Smouse (2001a, 2001b)there remain a number of key issues regarding the analysis of pollen pool genetic structure that have yet to be explored. Up to this point, we have focused on determining whether the means of the pollen pools sampled by each maternal individual are significantly different. In essence, both the TWOGENER and the Stepwise analyses test the hypothesis that the average pollen donor, or in other words, the centroid of the multivariate pollen pool, is the same across all mothers. From these results, the average dispersal distance and the genetic effective neighborhood size are easily estimated. However, variation in sampled pollen pools across mothers may be just as important. In terms of transferring genetic variation from one generation to the next, the differences in the variation observed within pollen pools is arguably more important than the mean differences among the means of the pollen pools. A method for testing the equality of variance, or heteroscedasticity, in sampled pollen pools could aid in identifying sites of high genetic diversity among the surrounding adults (RJ Dyer, in preparation).
Furthermore, these models have been constructed under the assumption of isotropic pollen movement (ie, pollen dispersal is equal in all directions). For coastal communities or populations in areas with predominant wind directions, directional pollen movement should be considered. Down-wind mothers will sample a significantly more diverse set of pollen donors than their upwind counterparts. This increase in diversity may lead to a significant increase in differentiation between down- and up-wind mothers, attributable entirely to the directionality of pollen dispersal. Similarly, we have consistently assumed, during the development of the TWOGENER analysis, that wind is the primary dispersal agent, restricting our inquiries to only a subset of plant taxa. Trap lining by insect or animal pollinators can significantly influence the heterogeneity of sampled pollen. At this time, we do not know how these behaviors would influence our interpretations of pollen movement, although there is active work in this area. The TWOGENER model has provided a means of testing hypotheses regarding the movement of pollen across landscapes. By extending the TWOGENER model (and underlying AMOVA analysis) to a generalized linear model, we gain the ability to investigate other factors affecting pollen pool differentiation and pollen dispersal.
References
Adams WT, Birkes DS (1991). Estimating mating patterns in forest tree populations. In: Fineschi S, Malvolti ME, Cannata F, Hattemer HH (eds) Biochemical Markers in the Population Genetics of Forest Trees. SPB Academic Publishing: The Hague, Netherlands pp 157–172.
Allard RW, Garcia P, Saenz-de-Miera LE, Peres de la Vega M (1993). Evolution of multilocus genetic structure in Avena hirtula and Avena barbata. Genetics 135: 1125–1139.
Austerlitz F, Smouse PE (2001a). Two-generation analysis of pollen flow across a landscape, II. Relation between ΦFT, pollen dispersal and inter-female distance. Genetics 157: 851–857.
Austerlitz F, Smouse PE (2001b). Two-generation analysis of pollen flow across a landscape. III. Impact of adult population structure. Genet Res 78: 271–280.
Dolgez A, Baril C, Joly HI (1998). Fine-scale spatial genetic structure with non-uniform distribution of individuals. Genetics 148: 905–919.
Draper NR, Smith H (1981). Applied Regression Analysis. John Wiley and Sons: New York.
Dyer RJ, Sork VL (2001). Pollen pool heterogeneity in shortleaf pine, Pinus echinata (Mill). Mol Ecol 10: 859–866.
Dyer RJ, Sork VL (2002). The effects of autocorrelated patterns among adults on pollen pool differentiation. In: Degan B, Loveless MD, Kremer A (eds) Modeling and Experimental Research on Genetic Processes in Tropical and Temperate Forests. LES COLLOQUES DE L’INRA, Kourou, French Guyana. pp 89–93.
Ellstrand NC, Elam DR (1993). Population genetics of small population size, implications for plant conservation. Annu Rev Ecol Syst 23: 217–242.
Excoffier L, Smouse PE, Quattro JM (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 479–491.
Farris MA, Mitton JB (1984). Population density, outcrossing rate, and heterozygote superiority in Ponderosa pine. Evolution 38: 1151–1154.
Gower JC (1966). Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53: 325–338.
Gram WK, Sork VL (1999). Population density as a predictor of genetic variation for woody plant species. Cons Bio 13: 1079–1087.
Gram WK, Sork VL (2001). Association between environmental and genetic heterogeneity in forest tree populations. Ecology 82: 2012–2021.
Harper JL (1977). Population biology of plants. Academic Press: Orlando Florida.
Henderson CR (1953). Estimation of variance and covariance components. Biometrics 9: 226–252.
Johnson RA, Wichern DW (1992). Applied Multivariate Statistical Analysis, Third Edition. Prentice-Hall: New Jersey.
Knowles P, Furnier GR, Aleksiuk MA, Perry DJ (1987). Significant levels of self-fertilization in natural populations of Tamarack. Can J Bot 65: 1087–1091.
Koop AL (1996). Genetic variation and structure in Quercus alba L. in a Missouri Ozark landscape. MS thesis. University of Missouri-Saint Louis.
Ledig FT (1992). Human impacts on genetic diversity in forest ecosystems. Oikos 63: 87–108.
Levin DA, Kerster HW (1974). Gene flow in seed plants. Evol Biol 7: 139–220.
Loechelt S, Franke A (1996). Genetic constitution of Beech stands (Fagus sylvatica L.). along an altitudinal transect from Freiburg to the top of ‘Mount Schauinsland’. Sil Genet 44: 312–318.
Murawski DA, Hamrick JL (1991). The effect of the density of flowering individuals on the mating systems of nine tropical tree species. Heredity 67: 141–167.
Okubo A, Levin SA (1989). A theoretical framework for data analysis of wind dispersal of seed and pollen. Ecology 70: 329–338.
Sampson JF, Hopper SD, James SH (1990). Temporal variation in allele frequencies in the pollen pool of Eucalyptus rhodantha. Heredity 65: 189–200.
Searle SR (1968). Another look at Henderson's methods of estimating variance components. Biometrics 24: 749–788.
Searle SR (1997). Linear Models, Wiley Classics Library Edition. John Wiley and Sons: New York.
Shea KL (1987). Effects of population structure and cone production on out-crossing rates in Engelmann Spruce and Subalpine Fir. Evolution 41: 124–136.
Smouse PE, Dyer RJ, Westfall RD, Sork VL (2001). Two-generation analysis of pollen flow across a landscape. I. Male gamete heterogeneity among females. Evolution 55: 260–271.
Zelen M (1968). Discussion of Searle [1968]. Biometrics 24: 779–780.
Acknowledgements
We thank JF Fernandez and two anonymous reviewers for insightful comments regarding this manuscript. RJD received support through NSF (NSF-Dissertation Research-0073242) and through funding awarded to VLS by the University of Missouri Research Board and UM-St Louis Research Award programs. VLS was additionally supported by NSF-DEB-0089445. PES was supported by the National Science Foundation (NSF-BSR-0089238 and NSF-BSR-0211430), the New Jersey Agricultural Experimental Station (NJAES-17109 and McIntire-Stennis 17309). Elements of this work were conducted as part of the Gene Flow Dynamics Working Group supported by the National Center for Ecological Analysis and Synthesis, a Center funded by NSF (Grant #DEB-0072909), the University of California, and the Santa Barbara campus.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dyer, R., Westfall, R., Sork, V. et al. Two-generation analysis of pollen flow across a landscape V: a stepwise approach for extracting factors contributing to pollen structure. Heredity 92, 204–211 (2004). https://doi.org/10.1038/sj.hdy.6800397
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800397
Keywords
This article is cited by
-
Pollen-mediated transfer of herbicide resistance between johnsongrass (Sorghum halepense) biotypes
Scientific Reports (2022)
-
Rangewide population differentiation and population substructure in Quercus rubra L.
Tree Genetics & Genomes (2017)
-
Genetic variation in natural populations of Acacia visco (Fabaceae) belonging to two sub-regions of Argentina using AFLP
Plant Systematics and Evolution (2016)
-
Pollination graphs: quantifying pollen pool covariance networks and the influence of intervening landscape on genetic connectivity in the North American understory tree, Cornus florida L.
Landscape Ecology (2012)
-
Landscape genetic structure of natural populations of Acacia caven in Argentina
Tree Genetics & Genomes (2012)