Abstract
The Chudley–Lowry syndrome (ChLS, MIM 309490) is an X-linked recessive condition characterized by moderate to severe mental retardation, short stature, mild obesity, hypogonadism, and distinctive facial features characterized by depressed nasal bridge, anteverted nares, inverted-V-shaped upper lip, and macrostomia. The original Chudley–Lowry family consists of three affected males in two generations. Linkage analysis had localized the gene to a large interval, Xp21–Xq26 and an obligate carrier was demonstrated to have highly skewed X inactivation. The combination of the clinical phenotype, consistent with that of the patients with ATR-X syndrome, the skewed X-inactivation pattern in a carrier female, as well as the mapping interval including band Xq13.3, prompted us to consider the XNP/ATR-X gene being involved in this syndrome. Using RT-PCR analysis, we screened the entire XNP/ATR-X gene and found a mutation in exon 2 (c.109C>T) giving rise to a stop codon at position 37 (p.R37X). Western blot and immunocytochemical analyses using a specific monoclonal antibody directed against XNP/ATR-X showed the protein to be present in lymphoblastoid cells from one affected male, despite the premature stop codon. To explain these discordant results, we further analyzed the 5′ region of the XNP/ATR-X gene and found three alternative transcripts, which differ in the presence or absence of exon 2, and the length of exon 1. Our data suggest that ChLS is allelic to the ATR-X syndrome with its less severe phenotype being due to the presence of some XNP/ATR-X protein.
Similar content being viewed by others
Introduction
The Chudley–Lowry syndrome (ChLS, MIM 309490) is an X-linked recessive condition characterized by moderate to severe mental retardation, short stature, mild obesity, hypogonadism, a low total finger ridge count, and a distinctive face characterized by bitemporal narrowness, almond-shaped palpebral fissures, depressed nasal bridge, anteverted nares, short inverted V-shaped upper lip, and macrostomia. The original family previously described by us had three affected males in two generations.1 Linkage analysis had localized the morbid gene to a large interval, Xp21–Xq26. Subsequently, an obligate carrier was demonstrated to have a highly skewed X-inactivation pattern (95:5) (FE Abidi and CE Schwartz, unpublished data).
The clinical signs observed in these patients, particularly the facial dysmorphism and the mental retardation, were suggestive of the ATR-X syndrome [MIM 301040].2,3 The gene involved in the ATR-X syndrome, named XNP/ATR-X, maps to Xq13.3 and contains both zinc-finger and helicase domains. This gene has been found to be responsible for several other syndromes such as Juberg–Marsidi,4 Carpenter–Waziri,5 Holmes–Gang,6 Smith–Fineman–Myers syndromes,7 and one form of XLMR with spastic paraplegia.8 All these syndromes, now designated as the XLMR-hypotonic facies syndromes,6 have characteristic facial dysmorphology, severe mental retardation, and a highly skewed X-inactivation pattern in obligate carrier women.8
All these data suggested the XNP/ATR-X gene as a good candidate for ChLS. To test this hypothesis, we performed direct sequencing of the entire coding region (7476 bp) of the XNP/ATR-X gene and identified a nonsense mutation in exon 2 resulting in a stop codon at position 37 (p.R37X). Moreover, using Western blot analysis, we found that the XNP/ATR-X protein was present in the lymphoblastoid cells isolated from the proband. Based on this result and published data describing an alternative splicing event involving exon 6, we looked for transcripts with different 5′-ends of the XNP/ATR-X gene.9,10 Here, we describe three alternatively spliced transcripts, which are different in the first two exons of the XNP/ATR-X gene and lead to potential proteins differing in their NH2-terminal regions. Two transcripts result from the utilization of different splice donor sites within exon 1 and the third consists of an alternative splicing out of the exon 2. The isoform lacking exon 2 or the utilization of a downstream initiation codon would explain the expression of some XNP/ATR-X protein in patients with ChLS, and the fact they are less severely affected than most ATR-X patients. Our findings confirm the hypothesis that the ChLS was allelic to the XLMR-hypotonic facies syndromes (ATR-X).2,6
Materials and methods
Patients
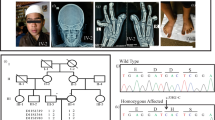
We previously described three males of the same family affected by the ChLS.1 The proband (Figure 1a, V-3) and two maternal uncles (IV-5, IV-6) were re-examined in 2002. All the affected males exhibited a clinical phenotype consistent with the XLMR-hypotonic facies syndrome such as short stature, small head circumference, open mouth, and inverted prominent upper lip [MIM 301040].3
Segregation analysis of the c.109C>T mutation in family K8937. The PCR product was generated using primers XNP-Ex2BF and XNP-Ex2BR, digested with NlaIII and resolved on a 2.5% agarose gel. (a) Partial pedigree of family K8937 is shown. The numbers below the obligate carrier females indicate the X-inactivation status. (b) The normal males have a 230 bp fragment, whereas the affected males have two fragments, 178 and 52 bp. The pattern of the obligate carriers consists of fragments of 230, 178, 52, and 12 bp. A 12 bp band, present in all individuals, is not seen on the gel.
X-inactivation studies
X-inactivation was determined by methylation analysis for the androgen receptor (AR) locus using a modification of the assay described by Allen et al.11. In all, 200 ng of genomic DNA from each patient was digested in a total volume of 10 μl with 5 U of Rsa1 alone or 5 U of Rsa1 and 10 U of HpaII (NEB, Beverly, MA, USA) for 16 h at 37°C. The enzymes were heat inactivated at 95°C for 10 min. A measure of 1 μl of each digested sample was amplified in a total volume of 20 μl containing 1 × PCR buffer (Sigma, St Louis, MO, USA), 100 μ M dNTPs, 1 μ M of each primers (AR-F: fluorescein amidite (Fam) 5′-gct gtgaaggttgctgttcctcat-3′; and AR-R: 5′-tccagaatctgttcca gagcgtgc-3′) and 1 U Taq polymerase (Sigma, St Louis, MO, USA). PCR conditions were as follows: 95°C for 5 min, then 28 cycles of 95°C for 30 s, 60°C for 30 s, 72°C for 30 s. After checking for amplification on a 2% agarose gel, the samples were run for quantitative analysis on the Automated Laser Fluorescence (ALF) DNA sequencer (Amersham Biosciences, Piscataway, NJ, USA). The forward primer was labeled with Fam, which allowed the quantitative analysis as described by Pegoraro et al.12
RT-PCR and sequence analysis
Total RNA was extracted from Epstein–Barr virus (EBV)-transformed lymphoblasts from the proband, using the GenEluteTM Mammalian total RNA kit (Sigma, St Louis, MO, USA).13 The total RNA was treated with 1 U of DNaseI (Invitrogen, Carlsbad, CA, USA) for 15 min at room temperature followed by a purification step using the RNAeasy Mini kit (QIAGEN, Valencia, CA, USA). Messenger RNA was extracted from the various sources using the QuickPrep mRNA purification kit, according to the instructions of the manufacturer (Amersham Biosciences, Piscataway, NJ, USA). Approximately 3–5 μg of total RNA or 500 ng of mRNA was used in the first-strand cDNA synthesis with random hexamer primers using the SuperScript first-strand synthesis system for RT-PCR (Invitrogen, Carlsbad, CA, USA). A  volume of template was used in 50 μl of PCR mixture, containing 10 μ M of each primer, 250 μ M of dNTPs, 1 × PCR buffer (10 mM Tris, 50 mM KCl, 1.5 mM MgCl2 (pH 8.3)) and 1.5 U Taq polymerase (Sigma, St Louis, MO, USA). PCR conditions were as follows: 95°C for 5 min, followed by 30–35 cycles of 95°C for 30 s, 55–60°C for 30 s, and 72°C for 50 s, followed by a final extension at 72°C for 7 min. In total, 15 sets of overlapping primer pairs were designed to analyze the entire ORF (7476 bp) of the XNP/ATR-X gene. Primers, ATRX-cDNA1F (5′-atcggcgcccagcaatcacaga-3′) and ATRX-cDNA8R (5′-tccaccttccgcacaccacctaca-3′) spanning exons 1–8 detected the alteration described in this paper. Primer sequences for the rest of the coding region are available upon request. Primers 5′H2 (5′-ggtgtcgctttgtttccgcg-3′) and 5′KR (5′-gacgatcctgaagacttggat-3′) were also used to look for the presence and expression of alternative transcripts corresponding to the XNP/ATR-X gene. RT-PCR products were run on an agarose gel to confirm fragment size followed by a purification step through QIAGEN columns. Products were sequenced in both directions on an ALF DNA sequencer (Amersham Biosciences, Piscataway, NJ, USA) using the Thermosequenase CyTM5 dye terminator kit (Amersham Biosciences, Piscataway, NJ, USA) according to the manufacturer's protocol. The DNASTAR program was used for alignment and analysis of the sequences.
volume of template was used in 50 μl of PCR mixture, containing 10 μ M of each primer, 250 μ M of dNTPs, 1 × PCR buffer (10 mM Tris, 50 mM KCl, 1.5 mM MgCl2 (pH 8.3)) and 1.5 U Taq polymerase (Sigma, St Louis, MO, USA). PCR conditions were as follows: 95°C for 5 min, followed by 30–35 cycles of 95°C for 30 s, 55–60°C for 30 s, and 72°C for 50 s, followed by a final extension at 72°C for 7 min. In total, 15 sets of overlapping primer pairs were designed to analyze the entire ORF (7476 bp) of the XNP/ATR-X gene. Primers, ATRX-cDNA1F (5′-atcggcgcccagcaatcacaga-3′) and ATRX-cDNA8R (5′-tccaccttccgcacaccacctaca-3′) spanning exons 1–8 detected the alteration described in this paper. Primer sequences for the rest of the coding region are available upon request. Primers 5′H2 (5′-ggtgtcgctttgtttccgcg-3′) and 5′KR (5′-gacgatcctgaagacttggat-3′) were also used to look for the presence and expression of alternative transcripts corresponding to the XNP/ATR-X gene. RT-PCR products were run on an agarose gel to confirm fragment size followed by a purification step through QIAGEN columns. Products were sequenced in both directions on an ALF DNA sequencer (Amersham Biosciences, Piscataway, NJ, USA) using the Thermosequenase CyTM5 dye terminator kit (Amersham Biosciences, Piscataway, NJ, USA) according to the manufacturer's protocol. The DNASTAR program was used for alignment and analysis of the sequences.
Mutation Analysis
The c.109C>T alteration created an NlaIII site. New primers, XNP-Ex2BF (5′-agaagcttcatgacttccttgcac-3′) and XNP-Ex2BR (5′-aggggcttctataaagcttgctaatct-3′), were designed to amplify the region spanning the alteration. The 242 bp product was then digested with NlaIII (NEB, Beverly, MA, USA) and the digestion products were run on a 2.5% agarose gel. The normal sequence yielded fragments of 12 and 230 bp, whereas the altered sequence resulted in fragments of 12, 52, and 178 bp. To rule out a rare polymorphism, 100 X chromosomes from normal males were tested and none of them had this alteration.
Protein extraction and Western blot analysis
Proteins were extracted from lymphoblastoid cell lines of a normal individual, the proband V-3, and an ATR-X patient (p.R246C) using a modified lysis buffer (50 mM Tris-HCl (pH=7.4), 100 mM NaCl, 0.4% NP-40, pepstatin 10 μg/ml, 10 mM β-mercaptoethanol, 50 mM NaF, 1 mM Na3VO4, 30 mM pyrophosphate, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitors) (CompleteMini, Roche, Indianapolis, IN, USA) by mixing for 5 min at 4°C. The extracts were centrifuged at 40 000 r.p.m. for 30 min at 4°C in an ultracentrifuge tube and the supernatant was frozen at −80°C. Protein expression was assessed by Western blot analysis. In brief, samples were boiled and resolved by SDS-PAGE on 6% gels, which were subsequently transferred to a PVDF Western blotting membrane (Roche, Indianapolis, IN, USA) at 10 V using the Novex Xcell II electrophoretic blotter (Invitrogen, Carlsbad, CA, USA) in a buffer of Tris-glycine/20% methanol. The membrane was blocked for 1 h in 5% or 10% skim milk powder in TBST (Tris-buffered saline with 0.05% Tween 20) at room temperature. The membranes were then incubated with the mouse monoclonal XNP antibody 2H12 (diluted at 1/100) for 1 h.14 HRP-conjugated secondary antibody was incubated with the blot for 45 min at room temperature. Enhanced chemiluminescence reagents were used for detection (ECL plus, Amersham Biosciences, Piscataway, NJ, USA).
Immunofluorescence staining
The cell preparations were obtained and treated as described previously.14 After fixation, the cells were permeabilized with 0.1% Triton X-100 in PBS for 15 min at room temperature. Nonspecific staining was blocked with 1% bovine serum albumin (BSA) in PBS for 1 h before immunofluorescence staining. The cell preparations were incubated with monoclonal XNP antibody 2H12 (dilution 1:100) for 1 h at room temperature.14 Following three consecutive 5 min washes in PBS, cells were incubated for a further 30 min with Cy3-conjugated Goat anti-mouse antibody at 1:40 dilution in PBS, 1% BSA. Slides were washed three times with PBS before counterstaining of nuclear DNA with DAPI stain (100 ng/ml) diluted in Vectashield (Vector laboratories, Burlingame, CA, USA). Preparations were observed under a fluorescence microscope (Zeiss Axioplan-2) and the images captured with a CCD camera (Photometric, Tuscon, AZ, USA). Information was collected and merged using IPLab spectrum software (Vysis, Downers Grove, IL, USA).
Results
ChLS is caused by a nonsense mutation within the XNP/ATR-X gene
Mutation analysis of the XNP/ATR-X gene was performed by direct sequencing of the genomic regions coding for the two functional domains of the protein, which contain 85% of the mutations identified so far.15 By this approach, we failed to find a mutation in either the zinc-finger or the helicase domains of the patient V-3 (data not shown). Therefore, we designed a RT-PCR strategy to analyze the entire coding region of the XNP/ATR-X gene using 15 sets of overlapping primers. Sequencing of the RT-PCR product spanning exons 1–8 revealed a C>T substitution at position 109 in exon 2, resulting in a protein truncation (p.R37X). This nonsense mutation created an NlaIII restriction enzyme site in the abnormal sequence, which made it possible to test for segregation of the mutation in the family. The p.R37X mutation was found to segregate with the disease (Figure 1b). Additionally, the proband's sister (V-4) and the two maternal aunts (IV-3, IV-7) were found to be carriers of the mutant allele (Figure 1). This truncating mutation was not observed in 100 X chromosomes from normal individuals.
Since the presence of an X-linked mutation may cause the presence of skewed X-chromosome inactivation in the lymphocytes of carrier women,16 we tested the X-inactivation profile in each woman of this family, using the AR gene polymorphic repeated sequence on DNA prepared from fresh blood samples. All the obligate carrier women exhibited a highly skewed X-inactivation pattern (Figure 1a), which is consistent with findings previously reported for families with XNP/ATR-X mutations.8
Mutation p.R37X does not completely abolish the expression of the XNP/ATR-X protein
In order to determine the level of expression of the XNP/ATR-X protein in the Chudley–Lowry patients carrying the p.R37X mutation, we performed Western blot analysis. A monoclonal antibody directed against the XNP/ATR-X protein was used on total cellular protein extracts from lymphoblastoid cell lines from a normal individual, the patient V-3, and an ATR-X patient carrying the missense mutation, p.R246C.14 The latter patient was used as a negative control, since the mutation led to the absence of expression of the XNP/ATR-X protein.14 A band of ∼280 kDa, attributable to XNP/ATR-X, was detected in the normal control as well as in the Chudley–Lowry patient but not in the ATR-X patient (Figure 2a). We confirmed these results by using immunofluorescence staining with the monoclonal anti-XNP antibody performed on cell preparations of the same subjects. Similar staining patterns are observed in cell nuclei from the normal individual (Figure 2b(1)) and the Chudley–Lowry patient (Figure 2b(2)). In contrast, cell nuclei from the ATR-X patient with the p.R246C missense mutation exhibited a diffuse XNP/ATR-X staining (Figure 2b(3)). Additionally, RT-PCR analysis demonstrated that the premature stop codon (p.R37X) does not affect the stability of the altered XNP transcript as amplification was achieved with all primer sets spanning the rest of the gene (data not shown).
Immunoblot analysis and cellular distribution of XNP/ATR-X protein in a control, the ChLS proband and an ATR-X patient. (a) Total protein extracts were obtained from lymphoblastoid cell lines of a normal individual, the proband V-3 (p.R37X), and an ATR-X patient (p.R246C) resolved by SDS-PAGE, transferred to nitrocellulose membrane and probed with an anti-XNP antibody. Proband V-3 (lane 3) showed expression of anXNP/ATR-X protein similar in size to a normal individual (lane 1). As previously shown, the ATR-X patient (lane 2) failed to express the XNP/ATR-X protein.14 (b) Lymphocytes from a normal individual, the proband V-3 (p.R37X), and an ATR-X patient (p.R246C) were immunolabeled with XNP 2H12 antibody (red signals). In the control (b1) and the proband (b2), XNP/ATR-X protein localized exclusively within the nucleus showing the normal pattern.14,28 By comparison, the distribution in the lymphocytes of the ATR-X patient (b3) revealed any or a very faint diffuse nuclear signal.
Exons 1 and 2 of the XNP/ATR-X gene are alternatively spliced
To explain how a truncating mutation in exon 2 can still result in the presence of XNP/ATR-X protein, we investigated in detail the 5′ region of the XNP/ATR-X transcript by RT-PCR analysis, using primers 5′H2 and 5′KR located in exons 1 and 4, respectively. This strategy revealed that apart from the normal transcript (417 bp), which contains the p.R37X nonsense mutation, two alternative transcripts of 304 and 513 bp were also expressed (Figure 3a). By direct sequencing of these two newly identified isoforms, we found that the smaller transcript (304 bp) lacks exon 2 and the larger transcript (513 bp) results from the utilization of a different splice site at the 3′-end of exon 1. The translation of each isoform is represented in Figure 3b.
(a) RT-PCR analysis was carried out on the RNA isolated from the lymphoblastoid cells of the proband and a normal male. The PCR reaction was carried out using primers 5′H2 (exon 1) and 5′KR (exon 4). Lane 1: 100 bp ladder (Invitrogen); lane 2: proband (V-3); lane 3: normal male. (b) Schematic representation of the 5′ alternative isoforms of the XNP/ATR-X transcript. The position of the two different splice sites at the 3′ boundary of exon 1 is indicated by an arrow. The different proteins potentially encoded are represented at the bottom of each transcript and the size of the predicted protein is listed. The initiation methionine is underlined and the position of exon–intron boundaries is indicated by vertical lines below the sequence. (c) Presence of different isoforms in the XNP/ATR-X transcript. RT-PCR analysis was performed on RNA isolated from human embryonic, fetal, and adult tissues. The PCR products were generated using primers 5′H2 and 5’KR and analyzed on a 2% agarose gel. In addition to the normal transcript (417 bp), which is the most abundant, two alternative transcripts were also detected: the smaller transcript (304 bp) lacking exon 2 and the larger transcript (513 bp) resulting from the utilization of a different splice site at the 3′-end of exon 1. Moreover, in some tissues such as fetal liver, an additional transcript (349 bp) missing 68 bp in exon 2 could be detected. MW is a molecular weight marker.
In order to define the tissue specificity of these alternative transcripts, we analyzed their expression by RT-PCR in RNA extracted from different tissues (brain, liver, kidney, fibroblasts, lymphocytes, and placenta) and developmental stages (fetal and adult). As shown in Figure 3c, the normal transcript is the most abundant one in each tissue tested and the transcript lacking exon 2 is present in all samples, but at a much lower level. In contrast, the larger transcript is expressed at a very low concentration in lymphocytes and adult liver and is not present in placenta and fetal liver (Figure 3c). Moreover, in some tissues such as in fetal liver (Figure 3c), an additional transcript (349 bp) missing 68 bp in exon 2 could be detected. This transcript was also detected in a human fetal brain 5′ stretch library (data not shown).
Discussion
The XNP/ATR-X gene was originally thought to be exclusively involved in the X-linked mental retardation syndrome associated with α-thalassemia (ATR-X), but it is becoming clear that it is associated with several other XLMR syndromes.6 Mutations in the XNP/ATR-X gene have been found in several X-linked mental retardation syndromes such as Carpenter–Waziri, Holmes–Gang, XLMR-spastic paraplegia, and Juberg–Marsidi, Smith–Fineman–Myers with various anomalies in multiple systems.3,4,5,6,7,8 In addition, Stevenson et al.6 proposed several other XLMR syndromes to be candidates for mutations in the XNP/ATR-X gene based on phenotypic similarities. Among these was the Chudley–Lowry, which has previously also been suggested to be allelic to ATR-X.2
In the present study, we have demonstrated that indeed the ChLS is allelic to the XLMR-hypotonic facies syndromes. The ChLS is caused by a base substitution, c.109C>T in the XNP/ATRX gene, which results in a nonsense mutation, p.R37X. This mutation should theoretically give rise to a severe phenotype due to the extremely early truncation of the protein. Surprisingly, immunocytochemical and Western blot analyses revealed the presence of XNP/ATR-X protein in the cells of the affected patient. This finding may account for the less severe phenotype observed in the ChLS.
An alternative splicing event, occurring in the 5′ region of the XNP/ATR-X transcript, could explain the presence of the XNP/ATR-X protein. Alternative splicing is now widely accepted as an important source of genetic diversity. To address this question, the 5′-end of the XNP/ATR-X transcript was analyzed in detail. We demonstrated the presence of an alternatively spliced form of the gene lacking exon 2. This transcript is apparently ubiquitously expressed albeit at a much lower level than the normal exon 2 containing transcript (Figure 3c). Since exon 2 contains the p.R37X mutation, the presence of the exon 2 minus transcript could account for the mild CLS phenotype. Recently, a conserved truncated isoform of the XNP/ATR-X protein, which lacks the SWI/SNF-homology domain has been identified.17 Identification of all XNP/ATR-X splice variants as well as elucidation of their functions may help to discover the exact role of this gene in various tissues.
Alternatively, an event other than alternative splicing could also explain the mild phenotype. The presence of an apparently stable transcript containing the c.109C>T alteration, which results in the p.R37X truncating mutation responsible, may reflect the utilization of a downstream methionine in exon 3, giving rise to a protein lacking exons 1 and 2.
These two alternative possibilities are consistent with the presence of a band at ∼280 kDa observed by Western blot analysis. The size is similar to the normal XNP/ATR-X protein. Alternative splicing or utilization of a downstream methionine will produce proteins differing from the normal protein by 4 or 6 kDa, respectively. Unfortunately, due to the size of the XNP/ATR-X protein it is not possible to determine which of these proteins is present on the Western blot based on size. This question requires utilization of more extensive protein analysis.
The p.R37X mutation has already been reported in a family with four patients affected by moderate to severe mental retardation and epilepsy.18 The comparison of clinical features observed in patients from both families showed mental retardation (moderate to severe) in all affected individuals and distinct facial features in all patients from the present family and in only two of four from the family reported by Guerrini et al.18 However, seizures were not observed in patients from the present family. In the family reported by Guerrini et al.18, only one patient was reported to be microcephalic, while in the present family, all three males had rather small head circumferences. This phenotypic variation suggests that the XNP/ATR-X protein, which belongs to the family of SWI/SNF DNA helicases, may influence the expression of several genes in multiple tissues during development. This hypothesis is supported by recent studies showing that the XNP/ATR-X protein is involved in a wide range of cellular functions including chromatin remodeling, regulation of the transcription,15,19 and functions not yet precisely defined. Thus, since the XNP/ATRX protein has multiple functional domains and likely interacts with many different proteins, variations in those proteins could explain the phenotype differences.
Our findings as well as those previously reported support the previous hypothesis that other XLMR syndromes are candidates for mutations in the XNP/ATR-X gene.4,5,6,7,20,21 Additionally, testing is needed to determine if syndromes like XLMR-Arch Fingerprints-Hypotonia, Brooks, Miles–Carpenter, Vasquez, and XLMR-Psoriasis may also be due to mutations in the XNP/ATR-X gene.22,23,24,25,26,27
Electronic-database information
The accession number and URLs for data presented herein is as follows: Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (Chudley–Lowry syndrome [MIM 309490], ATR-X [MIM 301040]). Genbank, http://www.ncbi.nlm.nih.gov/Genbank/ (for sequences for the XNP/ATR-X gene [accession number U75653])
References
Chudley AE, Lowry RB, Hoar DI : Mental retardation, distinct facial changes, short stature, obesity, and hypogonadism: a new X-linked mental retardation syndrome. Am J Med Genet 1988; 31: 741–751.
Cole TR, May A, Hughes HE : Alpha thalassaemia/mental retardation syndrome (non-deletional type): report of a family supporting X linked inheritance. J Med Genet 1991; 28: 734–737.
Stevenson RE, Schwartz CE, Schroer RJ : X-Linked Mental Retardation. New York: Oxford University Press Inc., 2000.
Villard L, Gecz J, Mattei JF et al: XNP mutation in a large family with Juberg–Marsidi syndrome. Nat Genet 1996; 12: 359–360.
Abidi F, Schwartz CE, Carpenter NJ, Villard L, Fontes M, Curtis M : Carpenter–Waziri syndrome results from a mutation in XNP. Am J Med Genet 1999; 85: 249–251.
Stevenson RE, Abidi F, Schwartz CE, Lubs HA, Holmes LB : Holmes–Gang syndrome is allelic with XLMR-hypotonic face syndrome. Am J Med Genet 2000; 94: 383–385.
Villard L, Fontes M, Ades LC, Gecz J : Identification of a mutation in the XNP/ATR-X gene in a family reported as Smith–Fineman–Myers syndrome. Am J Med Genet 2000; 91: 83–85.
Lossi AM, Millan JM, Villard L et al: Mutation of the XNP/ATR-X gene in a family with severe mental retardation, spastic paraplegia and skewed pattern of X inactivation: demonstration that the mutation is involved in the inactivation bias. Am J Hum Genet 1999; 65: 558–562.
Gibbons RJ, Bachoo S, Picketts DJ et al: Mutations in transcriptional regulator ATRX establish the functional significance of a PHD-like domain. Nat Genet 1997; 17: 146–148.
Villard L, Lossi AM, Cardoso C et al: Determination of the genomic structure of the XNP/ATRX gene encoding a potential zinc finger helicase. Genomics 1997; 43: 149–155.
Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW : Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet 1992; 51: 1229–1239.
Pegoraro E, Schimke RN, Arahata K et al: Detection of new paternal dystrophin gene mutations in isolated cases of dystrophinopathy in females. Am J Hum Genet 1994; 54: 989–1003.
Bolton BJ, Spurr NK : Culture of immortalized cells; in Freshney RI, Freshney MG (eds): B-Lymphocytes. New York: Weiley-Liss, 1996, pp 283–298.
Cardoso C, Lutz Y, Mignon C et al: ATR-X mutations cause impaired nuclear location and altered DNA binding properties of the XNP/ATR-X protein. J Med Genet 2000; 37: 746–751.
Villard L, Fontes M : Alpha-thalassemia/mental retardation syndrome, X-Linked (ATR-X, MIM #301040, ATR-X/XNP/XH2 gene MIM #300032). Eur J Hum Genet 2002; 10: 223–225.
Plenge RM, Stevenson RE, Lubs HA, Schwartz CE, Willard HF : Skewed X chromosome inactivation is a common feature of X-linked mental retardation disorders. Am J Hum Genet 2002; 71: 168–173.
Garrick D, Samara V, McDowell TL et al: A conserved truncated isoform of the ATR-X syndrome protein lacking the SWI/SNF-homology domain. Gene 2004; 326: 23–34.
Guerrini R, Shanahan JL, Carrozzo R, Bonanni P, Higgs DR, Gibbons RJ : A nonsense mutation of the ATRX gene causing mild mental retardation and epilepsy. Ann Neurol 2000; 47: 117–121.
Gibbons RJ, Higgs DR : Molecular–clinical spectrum of the ATR-X syndrome. Am J Med Genet 2000; 97: 204–212.
Villard L, Toutain A, Lossi AM et al: Splicing mutation in the ATR-X gene can lead to a dysmorphic mental retardation phenotype without alpha-thalassemia. Am J Hum Genet 1996; 58: 499–505.
Villard L, Lacombe D, Fontes M : A point mutation in the XNP gene, associated with an ATR-X phenotype without alpha-thalassemia. Eur J Hum Genet 1996; 4: 316–320.
Stevenson RE, Hane B, Arena JF et al: Arch fingerprints, hypotonia, and areflexia associated with X linked mental retardation. J Med Genet 1997; 34: 465–469.
Brooks SS, Wisniewski K, Brown WT : New X-linked mental retardation (XLMR) syndrome with distinct facial appearance and growth retardation. Am J Med Genet 1994; 51: 586–590.
Miles JH, Carpenter NJ : Unique X-linked mental retardation syndrome with fingertip arches and contractures linked to Xq21.31. Am J Med Genet 1991; 38: 215–223.
Proud VK, Levine C, Carpenter NJ : New X-linked syndrome with seizures, acquired microencephaly, and agenesis of the corpus callosum. Am J Med Genet 1992; 43: 458–466.
Vasquez SB, Hurst DL, Sotos JF : X-linked hypogonadism, gynecomastia, mental retardation, short stature, and obesity – a new syndrome. J Pediatr 1979; 94: 56–60.
Tranebjaerg L, Svejgaard A, Lykkesfeldt G : X-linked mental retardation associated with psoriasis: a new syndrome? Am J Med Genet 1988; 30: 263–273.
McDowell TL, Gibbons RJ, Sutherland H et al: Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc Natl Acad Sci USA 1999; 96: 13983–13988.
Acknowledgements
We would like to express our gratitude to the family for their participation in this study. We would like to thank C Skinner for coordinating sample collection. S Daniels provided sequencing support and T Moss was responsible for maintenance of tissue culture cell lines. This work was supported, in part, by a grant from NICHD (HD26202) to CES, a National Institute of Mental Health Grant (MH57840) to RES and a grant from the South Carolina Department of Disabilities and Special needs (SCDDSN). This work was also supported by a grant from the association pour la recherché contre le cancer (ARC) to MGM. This paper is dedicated to the memory of Ethan Francis Schwartz 1996–1998.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Abidi, F., Cardoso, C., Lossi, AM. et al. Mutation in the 5′ alternatively spliced region of the XNP/ATR-X gene causes Chudley–Lowry syndrome. Eur J Hum Genet 13, 176–183 (2005). https://doi.org/10.1038/sj.ejhg.5201303
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201303
Keywords
This article is cited by
-
The ATRX splicing variant c.21-1G>A is asymptomatic
Human Genome Variation (2022)
-
Targeting G-quadruplex DNA as cognitive function therapy for ATR-X syndrome
Nature Medicine (2018)
-
The role of genetics in the establishment and maintenance of the epigenome
Cellular and Molecular Life Sciences (2013)
-
Genetic syndromes caused by mutations in epigenetic genes
Human Genetics (2013)
-
Alpha-Thalassämie-Retardierungs-Syndrom
Monatsschrift Kinderheilkunde (2010)