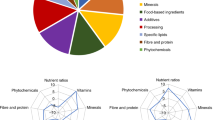
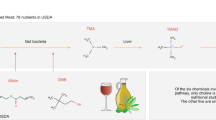
Abstract
Extensive programmes around the world endeavour to measure and catalogue the composition of food. Here we analyse the nutrient content of the full US food supply and show that the concentration of each nutrient follows a universal single-parameter scaling law that accurately captures the eight orders of magnitude in nutrient content variability. We show that the universality is rooted in the biochemical constraints obeyed by the metabolic pathways responsible for nutrient modulation, allowing us to confirm the empirically observed scaling law and to predict its variability in agreement with the data. We propose that the natural nutrient variability in food can be quantitatively formalized. This provides a mathematical rationale for imputing missing values in food composition databases and paves the way towards a quantitative understanding of the impact of food processing on nutrient balance and health effects.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The raw data are available at https://github.com/menicgiulia/FoodLaws. Source data are provided with this paper.
Code availability
The processing codes are available at https://github.com/menicgiulia/FoodLaws.
References
Kubo, R., Ichimura, H., Usui, T. & Hashitsume, N. Statistical Mechanics (North-Holland Personal Library, 1990).
Barabasi, A.-L. & Pósfai, M. Network Science by Albert-László Barabási (Cambridge University Press, 2016).
Barabási, A.-L., Menichetti, G. & Loscalzo, J. The nutritional dark matter: the unmapped chemical complexity of our diet. Nat. Food https://doi.org/10.1038/s43016-019-0005-1 (2019).
Hooton, F., Menichetti, G. & Barabási, A.-L. Exploring food contents in scientific literature with FoodMine. Sci. Rep. 10, 16191 (2020).
Milanlouei, S. et al. A systematic comprehensive lÿongitudinal evaluation of dietary factors associated with acute myocardial infarction and fatal coronary heart disease. Nat. Commun. 11, 6074 (2020).
Jeong, H., Tombor, B., Albert, R., Oltvai, Z. N. & Barabasi, A. L. The large-scale organization of metabolic networks. Nature 407, 651–654 (2000).
List of EuroFIR Databases (EuroFIR, accessed 7 January 2021); https://www.eurofir.org/food-information/food-composition-databases/
Placzek, S. et al. BRENDA in 2017: new perspectives and new tools in BRENDA. Nucleic Acids Res. 45, D380–D388 (2017).
USDA Food and Nutrient Database for Dietary Studies Version 5.0 (USDA, 2012); http://www.ars.usda.gov/ba/bhnrc/fsrg
Sebastian, R. et al. Flavonoid Values for USDA Survey Foods and Beverages 2007–2010 (USDA, 2016); www.ars.usda.gov/nea/bhnrc/fsrg
Willett, W. Monographs in Epidemiology and Biostatistics: Nutritional Epidemiology Vol. 15 (Oxford Univ. Press, 1990).
Hansen, A. The three extreme value distributions: an introductory review. Front. Phys. 8, 533 (2020).
Limpert, E., Stahel, W. A. & Abbt, M. Log-normal distributions across the sciences: keys and clues. Bioscience 51, 341–352 (2001).
FoodData Central (US Department of Agriculture, Agricultural Research Service, 2019); https://fdc.nal.usda.gov/
National Nutrient Database for Standard Reference, Release 28, Documentation and User Guide (USDA, 2015).
Frida Fooddata Version 2 (DTU Food, 2016).
Neveu, V. et al. Phenol-Explorer: an online comprehensive database on polyphenol contents in foods. Database (Oxford) 2010, bap024 (2010).
WishartLab (FooDB, 2017); http://foodb.ca/
FoodData Central: Foundation Foods (U.S. Department of Agriculture, A. R. S., 2019); https://fdc.nal.usda.gov/
Bar-Even, A., Noor, E., Flamholz, A., Buescher, J. M. & Milo, R. Hydrophobicity and charge shape cellular metabolite concentrations. PLoS Comput. Biol. 7, e1002166 (2011).
Muchowska, K.B., Varma, S.J. & Moran, J. Synthesis and breakdown of universal metabolic precursors promoted by iron. Nature 569, 104–107 (2019).
Chae, L., Kim, T., Nilo-Poyanco, R. & Rhee, S. Y. Genomic signatures of specialized metabolism in plants. Science 344, 510–513 (2014).
Park, J. O. et al. Metabolite concentrations, fluxes and free energies imply efficient enzyme usage. Nat. Chem. Biol. 12, 482–489 (2016).
Michal, G. & Schomburg, D. Biochemical Pathways: An Atlas of Biochemistry and Molecular Biology 2nd edn (John Wiley & Sons, 2013); https://doi.org/10.1002/9781118657072
Almaas, E., Kovács, B., Vicsek, T., Oltvai, Z. N. & Barabási, A.-L. Global organization of metabolic fluxes in the bacterium Escherichia coli. Nature 427, 839–843 (2004).
Almaas, E., Oltvai, Z. N. & Barabási, A. L. The activity reaction core and plasticity of metabolic networks. PLoS Comput. Biol. 1, 0557–0563 (2005).
Levine, E. & Hwa, T. Stochastic fluctuations in metabolic pathways. Proc. Natl Acad. Sci. USA 104, 9224–9229 (2007).
Stryer, L., Berg, M. J. & Tymoczko, L. J. Biochemistry (W. H. Freeman, 2002).
Peregrín-Alvarez, J.M., Sanford, C. & Parkinson, J. The conservation and evolutionary modularity of metabolism. Genome Biol 10, R63 (2009).
Khan, A. H., Zou, Z., Xiang, Y., Chen, S. & Tian, X. L. Conserved signaling pathways genetically associated with longevity across the species. Biochim. Biophys. Acta Mol. Basis Dis. 1865, 1745–1755 (2019).
Plant Metabolic Network (PlantCyc Pathway: Choline Biosynthesis, 2019); https://pmn.plantcyc.org/PLANT/NEW-IMAGE?type=PATHWAY&object=PWY-3385
Bulmer, A. M. G. On fitting the Poisson lognormal distribution to species-abundance data. Biometrics 30, 101–110 (1974).
Küken, A., Eloundou-Mbebi, J. M. O., Basler, G. & Nikoloski, Z. Cellular determinants of metabolite concentration ranges. PLoS Comput. Biol. 15, e1006687 (2019).
Bar-Even, A. et al. The moderately efficient enzyme: evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry 50, 4402–4410 (2011).
Dourado, H., Maurino, V. & Lercher, M. Enzymes and substrates are balanced at minimal combined mass concentration in vivo. Preprint at bioRxiv https://doi.org/10.1101/128009 (2017).
Vazquez, A. et al. Impact of the solvent capacity constraint on E. coli metabolism. BMC Syst. Biol. 2, 7 (2008).
Vazquez, A. Optimal cytoplasmatic density and flux balance model under macromolecular crowding effects. J. Theor. Biol. 264, 356–359 (2010).
Furusawa, C., Suzuki, T., Kashiwagi, A., Yomo, T. & Kaneko, K. Ubiquity of log-normal distributions in intra-cellular reaction dynamic. Biophysics (Nagoya-shi) 1, 25–31 (2005).
Beal, J. Biochemical complexity drives log-normal variation in genetic expression. Eng. Biol. 1, 55–60 (2017).
Salman, H. et al. Universal protein fluctuations in populations of microorganisms. Phys. Rev. Lett. 108, 238105 (2012).
Taniguchi, Y. et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329, 533–539 (2011).
Kærn, M., Elston, T. C., Blake, W. J. & Collins, J. J. Stochasticity in gene expression: from theories to phenotypes. Nat. Rev. Genet. 6, 451–464 (2005).
Banavar, J. R., Maritan, A. & Rinaldo, A. Size and form in efficient transportation networks. Nature 399, 130–132 (1999).
Maritan, A., Rigon, R., Banavar, J. R. & Rinaldo, A. Network allometry. Geophys. Res. Lett. 29, 1508 (2002).
Enquist, B. J., Brown, J. H. & West, G. B. Allometric scaling of plant energetics and population density. Nature 395, 163–165 (1998).
Gallos, L. K., Song, C., Havlin, S. & Makse, H. A. Scaling theory of transport in complex biological networks. Proc. Natl Acad. Sci. USA 104, 7746–7751 (2007).
Cordain, L. et al. Origins and evolution of the Western diet: health implications for the 21st century. Am. J. Clin. Nutr. 81, 341–354 (2005).
Micha, R., Wallace, S. K. & Mozaffarian, D. Red and processed meat consumption and risk of incident coronary heart disease, stroke, and diabetes mellitus: a systematic review and meta-analysis. Circulation 121, 2271–2283 (2010).
Fiolet, T. et al. Consumption of ultra-processed foods and cancer risk: results from NutriNet-Sant‚ prospective cohort. Brit. Med. J. 360, k322 (2018).
Adjibade, M. et al. Prospective association between ultra-processed food consumption and incident depressive symptoms in the French NutriNet-Sant‚ cohort. BMC Med. 17, 78 (2019).
Alonso-Pedrero, L. et al. Ultra-processed food consumption and the risk of short telomeres in an elderly population of the Seguimiento Universidad de Navarra (SUN) Project. Am. J. Clin. Nutr. 111, 1259–1266 (2020).
Carrera-Bastos, P., Fontes-Villalba, M., O'Keefe, J. H., Lindeberg, S. & Cordain, L. The western diet and lifestyle and diseases of civilization. Res. Rep. Clin. Cardiol. 2, 15–35 (2011).
Bornholdt, S. & Sneppen, K. Robustness as an evolutionary principle. Proc. R. Soc. Lond. B 267, 2281–2286 (2000).
Riehl, W. J., Krapivsky, P. L., Redner, S. & SegrŠ, D. Signatures of arithmetic simplicity in metabolic network architecture. PLoS Comput. Biol. 6, e1000725 (2010).
Segré, D., Shenhav, B., Kafri, R. & Lancet, D. The molecular roots of compositional inheritance. J. Theor. Biol. 213, 481–491 (2001).
Palsson, B. Systems Biology: Properties of Reconstructed Networks (Cambridge Univ. Press, 2006); https://doi.org/10.1017/CBO9780511790515
Gupta, S., Hawk, T., Aggarwal, A. & Drewnowski, A. Characterizing ultra-processed foods by energy density, nutrient density, and cost. Front. Nutr. 6 (2019).
Menichetti, G., Ravandi, B., Mozaffarian, D. & Barabasi, A.-L. Machine learning prediction of food processing. Preprint at medRxiv https://doi.org/10.1101/2021.05.22.21257615 (2021).
FNDDS Web Page (USDA, 2019); https://www.ars.usda.gov/northeast-area/beltsville-md-bhnrc/beltsville-human-nutrition-research-center/food-surveys-research-group/docs/fndds/
Kapur, J. N. Maximum-entropy Models in Science and Engineering (India, Wiley, 1989).
NCBI Taxonomy (National Center for Biotechnology Information, 2019); https://www.ncbi.nlm.nih.gov/taxonomy
Huerta-Cepas, J., Serra, F. & Bork, P. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638 (2016).
Yannai, S. Dictionary of Food Compounds with CD-ROM Choice Reviews Online Vol. 51 (Taylor & Francis, 2013).
Acknowledgements
This work was partially supported by NIH grant no. 1P01HL132825, American Heart Association grant no. 151708, ERC grant no. 810115-DYNASET and Rockefeller Foundation grant no. 2019 FOD 026. We thank J. Loscalzo for useful discussions and insights on enzyme kinetics, as well as M. Sebek and S. Ofaim for helping with the chemical classification and disambiguation.
Author information
Authors and Affiliations
Contributions
G.M. and A.-L.B. conceived the project and wrote the manuscript. G.M. performed the data query, data integration, statistical analysis and analytical calculations.
Corresponding author
Ethics declarations
Competing interests
A.-L.B. is the founder of Scipher Medicine and Naring Health, companies that explore the use of network-based tools in health, and Datapolis, which focuses on urban data.
Peer review
Peer review information
Nature Food thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Sections 1–11, Figs. 1–22 and Tables 1 and 2.
Source data
Source Data Fig. 1
Nutrient values for PDFs and scaling laws.
Source Data Fig. 2
Extensive nutrient values for PDFs and scaling laws.
Source Data Fig. 3
KM values for PDFs and scaling laws.
Rights and permissions
About this article
Cite this article
Menichetti, G., Barabási, AL. Nutrient concentrations in food display universal behaviour. Nat Food 3, 375–382 (2022). https://doi.org/10.1038/s43016-022-00511-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43016-022-00511-0
This article is cited by
-
Improving the generalizability of protein-ligand binding predictions with AI-Bind
Nature Communications (2023)
-
Nutritional redundancy in the human diet and its application in phenotype association studies
Nature Communications (2023)
-
Machine learning prediction of the degree of food processing
Nature Communications (2023)
-
Improvement of nutritional quality of food crops with fertilizer: a global meta-analysis
Agronomy for Sustainable Development (2023)