Abstract
In quantum gas microscopy experiments, reconstructing the site-resolved lattice occupation with high fidelity is essential for the accurate extraction of physical observables. For short interatomic separations and limited signal-to-noise ratio, this task becomes increasingly challenging. Common methods rapidly decline in performance as the lattice spacing is decreased below half the imaging resolution. Here, we present an algorithm based on deep convolutional neural networks to reconstruct the site-resolved lattice occupation with high fidelity. The algorithm can be directly trained in an unsupervised fashion with experimental fluorescence images and allows for a fast reconstruction of large images containing several thousand lattice sites. We benchmark its performance using a quantum gas microscope with cesium atoms that utilizes short-spaced optical lattices with lattice constant 383.5 nm and a typical Rayleigh resolution of 850 nm. We obtain promising reconstruction fidelities ≳ 96% across all fillings based on a statistical analysis. We anticipate this algorithm to enable novel experiments with shorter lattice spacing, boost the readout fidelity and speed of lower-resolution imaging systems, and furthermore find application in related experiments such as trapped ions.
Similar content being viewed by others
Introduction
Efficient data processing based on machine learning techniques has found numerous applications ranging from pattern recognition to the classification of quantum many-body phases1,2. The potential of machine learning algorithms for applications in experimental quantum physics lies in its power to extract information from experimental or numerical data by reducing the available information to a few essential characteristics. Examples include the classification of topological phases of matter based on a limited number of experimental observables3,4, investigations of the phase diagram of the Fermi-Hubbard model based on (spin-resolved) density snapshots5,6 or the multi-parameter optimization of experimental cooling techniques for trapping and imaging of atoms7,8. These examples highlight the potential of machine learning techniques for faster and more accurate data analysis, in particular in the presence of noise or experimental imperfections. Machine learning has recently been considered theoretically in the context of quantum gas microscopes in order to relax the stringent experimental requirements for high-fidelity imaging9. In this work, we demonstrate a deep-learning architecture for reconstructing the optical lattice occupation from fluorescence images and benchmark it using experimental data.
Quantum gas microscopy has facilitated unprecedented levels of control and observation for the study of quantum many-body systems based on neutral atoms in optical lattices10,11,12. Experimentally, the system is typically probed using fluorescence imaging, where the atoms are pinned in deep optical lattices and scatter fluorescence photons which are recorded on a camera through a high-resolution imaging system. In order to extract complex observables such as counting statistics and (multi-point) correlation functions13, the lattice occupation needs to be reconstructed in a site-resolved fashion. The attainable accuracy of an observable thus directly depends on the accuracy of the reconstruction. The latter is fundamentally limited by two quantities: (1) The signal-to-noise ratio (SNR) of the recorded image and (2) the ratio β between the imaging resolution and the lattice spacing. Defining the resolution according to the Rayleigh criterion, most existing quantum gas microscopes work with resolutions that are close to the lattice spacing or up to a factor of 1.5 worse11,12,14,15,16,17,18,19,20,21,22,23.
In the past, several techniques have been developed to enable high-fidelity reconstruction: One of the first microscope experiments employed an iterative least-squares-based approach which is, however, computationally expensive and limited in fidelity for smaller SNR12,24. Deconvolution with a linear kernel that is constructed to have minimum overlap with atoms on adjacent sites in order to cancel the signal spillover25, on the other hand, is computationally fast, but limited in performance as it assumes a single kernel that does not depend on the number of neighboring atoms. However, especially for shorter lattice spacings, where the small interatomic separation enhances density-dependent effects during imaging such as superradiance, this assumption is violated26. Further approaches are based on image restoration techniques such as Wiener filtering27 or Richardson–Lucy deconvolution17,28,29. Both methods, however, rapidly decrease in fidelity for β ≳ 224. Moreover, unconstrained deconvolution methods can be improved by adding further information, such as the discrete lattice grid and a sophisticated noise model, as shown in ref. 30 for a one-dimensional (1D) system.
In general, all these algorithms aim to invert the convolution of the atomic distribution with the point spread function (PSF) of the imaging system, i.e., to realize a deconvolution, which is, however, ill-conditioned in the presence of experimental noise31. It has recently been recognized that neural networks can have advantages in solving such inverse problems due to their ability to approximate nonlinear relationships32,33. In addition, their low computational complexity promises a fast evaluation, especially compared to iterative algorithms. With respect to quantum gas microscopy, a reconstruction approach based on supervised neural networks has previously been proposed and benchmarked with simulated data9. The supervised nature, however, requires training using simulated fluorescence images. Hence, the performance in reconstructing experimental data will ultimately be limited by the accuracy of the simulation.
In this paper, we present an unsupervised deep-learning algorithm for the reconstruction of the lattice occupation from fluorescence images. The unsupervised nature allows us to train the network directly with experimental data, avoiding the need for any simulated training data. We experimentally benchmark the fidelity by analyzing the deconvolved count distributions as well as repeated exposures with data produced by our cesium quantum gas microscope that operates at a rather large resolution-to-spacing ratio of β = 2.2 (Fig. 1c). In particular, we find reconstruction fidelities ≳96% across all fillings, and we are able to reconstruct large images containing several thousand lattice sites in less than one second. Our scheme enables high-fidelity reconstruction at shorter lattice spacing compared to previous reconstruction algorithms. Besides potentially reducing the technical complexity of the imaging system, this is also important for special lattice configurations such as superlattices, triangular34,35,36, or state-dependent lattices37, as well as experiments working with dipolar interactions where close atomic distances are favorable38,39.
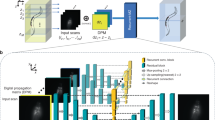
a Regularized convolutional autoencoder architecture, consisting of an encoder and a decoder. The input data passed to the encoder is a lattice section containing 16 × 16 lattice sites (256 × 256 pixels, white dots mark the lattice sites). This data is transformed by a sequence of five convolution layers, where in each layer, a discrete convolution with a set of learned kernels, is applied to the respective input. The third dimension in the output shape denotes the number of distinct kernels utilized per layer. Using a step size (stride) of two when advancing the kernels through the input during the convolution allows to reduce the image size by a factor of two in each step. At the end of the encoder, the node values of the bottleneck layer, after applying a tanh activation function, represent the binarized lattice occupancy. The node values before binarization contain the deconvolved counts in each lattice site, which exhibit a bimodal distribution owing to the saturating effect of the tanh function. Subsequently, the occupation matrix is processed by the decoder with the goal of replicating the input image. The decoder network transforms and upsamples the bottleneck layer using four transposed convolution layers. b Rectified Linear Unit (ReLU) activation functions are used in all layers except in the bottleneck and the last decoder layer, which use a tanh activation function. c Measured experimental point spread function (PSF, averaged over around 2000 individual PSFs) overlaid with the lattice grid. The central peak spans several sites and we observe long-range asymmetric features extending over the whole region of 16 × 16 lattice sites. d Signal-to-noise ratio determined from the count distributions of crops (1.6 μm crop width) containing exactly zero atoms or one atom. A background image without atoms was subtracted to shift the center of the background peak closer to zero.
Results
Network architecture
The problem at hand—reconstructing the binary lattice site occupation from a recorded fluorescence image —can be understood in the framework of data analysis as a problem of dimensionality reduction. Here, a high-dimensional noisy input image is transformed into the underlying binary lattice occupation of strongly reduced dimensionality. There exist a variety of dimensionality reduction techniques, with both linear approaches such as principal component analysis (PCA) as well as nonlinear methods40. In this work, we leverage the power of artificial neural networks, which fall into the latter category. Their inherent nonlinearity allows them to efficiently approximate the deconvolution32 as well as to capture density-dependent effects such as superradiance12,26. Among neural networks, the autoencoder is a popular architecture for various dimensionality reduction tasks41,42. Autoencoders consist of a stacked encoder and decoder network, separated by an interface layer that provides an encoded representation of the input data. The combined network is optimized to replicate the input data, enabling unsupervised training. We restrict the interface layer to a significantly lower dimensionality than the input, forcing the network to learn to extract the salient features of the input data in order to allow information flow through this bottleneck (Fig. 1a). In our context, the encoder learns the deconvolution from fluorescence image to site occupation, while the decoder learns to simulate a fluorescence image corresponding to this occupation.
We design and implement a regularized convolutional autoencoder architecture that is tailored to our reconstruction task. The network topology is depicted in Fig. 1a. Beginning from the left, the input layer takes a lattice section containing 16 × 16 lattice sites, corresponding to 256 × 256 pixels. The input image is then subsequently down-sampled by a set of four convolution layers (step size two, ReLU activation, Fig. 1b), before a final convolution operation (step size one, tanh activation, Fig. 1b) implements the bottleneck layer with an output matrix of 16 × 16 entries. This concludes the encoder part of the network, whose task is to reduce the input image to an array of site occupations. In the decoder part, the bottleneck layer is up-sampled by three transposed convolution layers (step size two, ReLu activation) and a final transposed convolution layer (step size one, tanh activation) to arrive at one output matrix with the same size as the input image.
In each of these layers, the input data is traversed by a set of kernels with entries learned during training, locally performing scalar multiplication (see Supplementary Note 2 for details on the convolution operations). After this discrete convolution, a nonlinear activation function is applied, producing one output for each kernel. The size of the output in the down-/up-sampling layers is always a factor of two lower/higher than the input resulting from the step size of two. An important aspect of our autoencoder implementation is that it consists exclusively of such convolution layers, which is required to retain spatial order throughout the network. Additionally, we split both the encoder and decoder into several convolution layers. Compared to a single convolution layer with kernels spanning the whole input, it was shown that deeper networks with smaller kernels perform significantly better in related applications43,44,45,46. While kernels in the first layer act directly on the raw image, subsequent layers are able to learn more complex, high-level features by combining the outputs of the previous layer. Additionally, the stride of two continuously changes the receptive field of kernels such that each layer is sensitive to correlations on different length scales47.
The final operation for determining the site occupancy in the bottleneck layer is the application of the activation function. Considering its sigmoidal shape (Fig. 1b), a truthful reconstruction of the occupation requires that the input values corresponding to empty and occupied sites saturate the activation function on opposite extremes, respectively. Therefore, the deconvolution implemented by the encoder must lead to a bimodal distribution of count values before binarization. The overlap between the two distributions is a signature of the quality of the reconstruction, which in principle, further enables a quantitative estimation of the reconstruction fidelity if the functional form of the two modes is known, as we will detail below.
The network is trained using a composite loss function
where x and \({{{{{{{\bf{{x}}}}}}}^{{\prime} }}}\) are the input and output images, respectively, yi are the node values in the bottleneck layer and Nsites is the number of sites in the image (here, Nsites = 162). The reconstruction loss \({{{{{{{{\mathcal{L}}}}}}}}}_{{{{{{{{\rm{L1}}}}}}}}}\left({{{{{{{\bf{x}}}}}}}},{{{{{{{\bf{{x}}}}}}}^{{\prime} }}}\right)={\sum }_{{{{{\rm{pixel}}}}}}| {{{{{{{\bf{x}}}}}}}}-{{{{{{{\bf{{x}}}}}}}^{{\prime} }}}|\) is augmented by an additional bottleneck regularization loss, where the regularization strength λ determines the relative weight between the two terms. The regularization term \({{N}_{{{{{{{{\rm{sites}}}}}}}}}}^{-1}\mathop{\sum }\nolimits_{i = 1}^{{N}_{{{{{{{{\rm{sites}}}}}}}}}}\left(1-| {y}_{i}| \right)\) penalizes non-binary values in the bottleneck layer, forcing the network to learn a transformation from the input image to the site occupation. Without the bottleneck regularizer (λ = 0), the network would learn to replicate the input image, but the bottleneck would contain a reduced representation of the input image with properties that are difficult to interpret. Our chosen form is one of several possible relations to promote binary node values48.
Training and application to experimental data
We benchmark the proposed reconstruction algorithm using experimental data. To this end, we prepare a single layer of ultracold cesium atoms in a two-dimensional optical lattice and image the atoms by scattering fluorescence photons on the D2 line with a high-resolution microscope (see Methods and Supplementary Note 1)49,50,51. The minimal distance between atoms during imaging is set by the pinning lattice, which has a spacing of a = 383.5 nm and the typical resolution according to the Rayleigh criterion is about 850 nm. The corresponding resolution-to-spacing ratio is β = 2.2, setting very challenging conditions for the reconstruction. Achieving a high reconstruction fidelity despite the short lattice spacing requires working at a comparably high SNR. Using the definition SNR = (catom − cbg)/(σatom + σbg), where catom,bg is the mean and σatom,bg the standard deviation of the count distribution for atoms and background, respectively, we find an experimental SNR of 5.2 for an exposure time of 300 ms (Fig. 1d).
In preparation for the reconstruction, we extract the lattice vectors from the Fourier transform of the positions of around 20 dilute atom images. The absolute position of the lattice phase with respect to the image origin is determined for each image by fitting the positions of a few isolated atoms (see Methods and Supplementary Note 2). Together, this fully defines the lattice grid and ensures that the sites are consistently located at the same positions within the local crops, spanning 16 × 16 lattice sites. During the training, the network learns that atoms can only be located at this discrete set of positions, which is important additional information to overcome the resolution limit.
We train the network using an experimental dataset of around 100,000 crops extracted from homogeneous clouds of various average fillings between zero and n ≈ 0.98 (see Methods and Supplementary Note 1 for information on how this data is obtained). The training procedure takes about 100 epochs (full passes through the training set) using the ADAM optimizer (initial learning rate 4 × 10−4)52. We performed hyperparameter tuning for the kernel sizes of the encoder and decoder, as well as the regularization strength λ. The latter strongly influences the performance of the network, which we optimize using the final training loss as well as the separation in the bimodal distribution of the deconvolved site counts. According to this, we determined an optimal value of λopt = 0.4. Additionally, we found optimal kernel sizes of 10 × 10 for the encoder and 22 × 22 for the decoder, respectively. Note, however, that beyond some minimal kernel size of about 8 × 8, the exact choice does not influence the performance of the network significantly. After successful training, we only use the encoder section of the network for reconstruction.
The reconstruction process is illustrated in Fig. 2, using an example image with a total size of 70 × 70 lattice sites. The raw image (Fig. 2a) is first decomposed into crops containing 16 × 16 sites. We advance only by one lattice site between each crop region, such that every site occurs in several analyses at different locations. Each crop is then fed into the encoder part of the network, where we extract the output of the bottleneck layer before binarization via the activation function. The encoder performs the learned deconvolution and transforms the input to a 16 × 16 matrix of deconvolved site counts. These matrices are finally reassembled according to their position in the original image, averaging the deconvolved counts across overlapping sites (Fig. 2b). During the assembly, we only take the central 12 × 12 sites of each crop into account as the border region suffers from a reduced fidelity due to edge effects as a result of missing neighboring sites that are important for the deconvolution. We find that the deconvolution results in a strong enhancement of the contrast of individual holes in the high-density plateau and vice versa of isolated atoms in the outside region. A histogram of all deconvolved site counts (Fig. 2c) reveals a bimodal distribution with two well-separated peaks, which we identify as empty and occupied sites, respectively. Since the tanh activation function that is used in the bottleneck layer is symmetric around zero, the correct discrimination threshold is, in general, zero as well. Applying this threshold to the deconvolved site count matrix finally gives the reconstructed occupation (Fig. 2d).
a Raw fluorescence image showing a Mott-insulating state (~2500 atoms) in a box potential (arrows mark the lattice orientation). Crops containing 16 × 16 sites are extracted from the full image and fed into the encoder part. b The deconvolved site counts are extracted from the bottleneck layer before binarization and reassembled according to their position in the full image. c A histogram of these values reveals a bimodal distribution with a clear separation between empty and occupied lattices sites, allowing to set a threshold for the site occupation. d Applying this threshold finally gives the reconstructed lattice occupation corresponding to the input image. Light gray circles denote empty and dark purple occupied sites.
A key challenge in employing artificial neural networks for data analysis is to ensure that the network learns a robust, physically reasonable transformation that generalizes to unseen data instead of memorizing the training samples or focusing on coincidental correlations. The symmetric structure of our network enables us to gain more insight into the learned behavior by isolating the decoder part and examining the generated output for varying binary input occupation matrices. We start by setting a single entry of the 16 × 16 occupation matrix to one, expecting that a single PSF appears at the appropriate location in the image generated by the decoder. Figure 3a, b show a comparison between the learned PSF and the measured one, which was extracted from many dilute images (similar to Fig. 1c). Both the cross-section and the 2D images visualize that the network learns a PSF matching the experimental one in size and shape, with the only difference between them being a small overall offset. As a second test, we investigate to what extent the network is able to approximate density-dependent effects, which are present in our experimental data. To this end, we generate an occupation matrix representing a block of occupied sites (Fig. 3c). We find that the resulting output image from the decoder is significantly brighter than a simple convolution of the occupation matrix with the measured PSF. To study this quantitatively, we generate random 16 × 16 occupation matrices at various filling fractions and process them using the decoder network. The output is compared to a convolution of the occupation matrix with the measured PSF as well as crops from experimental data (12 × 12 sites crop size) at the same mean filling. Plotting the mean counts as a function of the filling (Fig. 3d) reveals a significantly increased brightness for higher fillings in the decoder-generated images, which is compatible with the experimental data. This implies that the network is indeed able to capture the density-dependent effects due to superradiance, which in our experiment leads to a 22% higher signal at unity filling.
The transformations learned by the network can be visualized by extracting the decoder part and feeding different occupation matrices into the bottleneck layer. a Analysis of a single occupied site: comparison of the decoder output to the measured point spread function (PSF, averaged over around 2000 individual PSFs). b Cut through the center of the 2D images shown in a. c Analysis of a group of 9 × 9 occupied sites: Compared to a convolution with the measured PSF, the decoder output yields a significantly brighter image. d Mean intensity as a function of filling in regions cropped from experimental data (gray triangles), the decoder output for random occupation matrices at a given filling (purple circles) and a convolution of the same occupation with the measured PSF shown in a (orange diamonds). We only use the central 12 × 12 sites of the crops to avoid edge effects, and the experimental data points are computed from ~1500 crops per filling bin (same crop size). The lines are linear fits through the respective data points and the error bars denote the standard deviation.
Performance evaluation
In an experimental setting, the reconstruction fidelity is limited by the finite SNR, systematic errors such as spatially inhomogeneous fluorescence as well as hopping and atom loss. A direct measurement of the fidelity in the presence of these effects is hindered by the imperfect knowledge of the occupation labels in experimental images. One option would be to benchmark the algorithm on simulated data, but conceiving a simulation that accurately captures all effects present in the experiment is a highly challenging task and beyond the scope of this work. Instead, we present two approaches that allow us to estimate the experimental fidelity directly from the reconstruction process applied to experimental images.
First, we analyze the distribution of deconvolved site counts by extracting the node values of the bottleneck layer before binarization. Figure 4 shows the extracted site count distributions for four selected average fillings, where the value for the filling is computed from the reconstruction. We find well-discriminated bimodal distributions across all fillings, with a vanishing overlap at low filling (upper left panel), and a slightly increased overlap towards higher fillings. Without any processing, the distributions of empty and occupied sites would completely overlap due to imaging noise and the spillover of neighboring sites (see Supplementary Note 2). The effect of the deconvolution is now to separate these distributions again, which is possible up to a certain limit given by the aforementioned experimental imperfections. The remaining overlap can then be used as an approximation to estimate the reconstruction fidelity. In principle, it is expected that the fidelity is maximal for fillings close to zero and unity, and drops to a minimum around half-filling due to the maximum possible number of nearest and next-nearest neighbor configurations. In addition, density-dependent effects such as superradiance and the increased effective atom loss due to hopping can reduce the contrast of holes in experiments at high filling.
The deconvolved site counts are extracted from the bottleneck layer before binarization via the activation function. A bimodal distribution distinguishing between empty and occupied sites is found across all fillings. The overlap vanishes for small fillings and increases slightly towards higher fillings. To quantify the fidelity from the overlap, we fit two Gaussians to the data for half-filling (solid line), capturing the distributions of empty (dotted line, left peak) and occupied (dashed line, right peak) sites, respectively. Insets show a zoom-in of the overlap region. The filling values were obtained directly from the reconstruction using a threshold of zero, and the occurrences are normalized to the respective maximum in each plot. Each histogram is computed from ~1500 crops with 12 × 12 sites.
A drawback of the nonlinear transformation applied by the encoder is that the resulting functional form of the bimodal distribution is a priori not known. This makes it challenging to accurately quantify the reconstruction fidelity. Nevertheless, the deconvolved counts for the case of half-filling appear to be well described by a bimodal normal distribution. We can therefore estimate the fidelity of the reconstruction process from the overlap region between the two peaks by fitting two Gaussians to the deconvolved counts at half-filling (upper right panel). The overlap area corresponds to wrongly classified sites when discriminating via a threshold value, and the reconstruction fidelity is given by the fraction of this overlap area to the total area under the bimodal distribution. From the fit, we determine an experimental reconstruction fidelity for half-filling of \({{{{{{{\mathcal{F}}}}}}}} \sim 99 \%\).
Second, we quantify the reconstruction fidelity by taking two consecutive images of an atomic sample in the same experimental realization and comparing the reconstruction results. Such a double-imaging protocol has previously been used to quantify the probability of losses and hopping during imaging when assuming a perfect reconstruction12,14,16,18. However, as we will detail below, this can also be used to estimate the reconstruction fidelity in the presence of both finite hopping and losses during imaging as well as reconstruction errors. This method is sensitive to statistical fluctuations of the recorded counts at finite SNR, which limits the overall detection fidelity. We can derive a relation for the reconstruction fidelity \({{{{{{{\mathcal{F}}}}}}}}\):
where δ is the probability of finding different reconstruction results on a lattice site between the two images and pδ accounts for thermal hopping and atom loss, which was calibrated independently (see Supplementary Note 4 for a detailed derivation).
Figure 5 shows the measured reconstruction fidelity \({{{{{{{\mathcal{F}}}}}}}}\) as a function of the average filling n. We show data points both with (pδ(n) = n ⋅ 5.9 × 10−3) and without (pδ = 0) the hopping and loss correction. We find a reconstruction fidelity above 99% close to unity filling, and for n ≲ 0.2. A minimum occurs around n = 0.7, yielding an uncorrected reconstruction fidelity of 96.3(3)%.
By measuring the same many-body state twice, we can estimate the reconstruction fidelity from the difference in the reconstructed occupations via Eq. (2). The uncorrected data is obtained by neglecting hopping and loss, i.e., pδ = 0, while the corrected points (shaded) take the independently calibrated value pδ(n) = n ⋅ 5.9 × 10−3 into account. The zoom-in panels show two typical images at the indicated mean filling with the respective first and second exposure. The blue squares in the second image indicate sites that changed from unoccupied to occupied, and the green crosses vice versa. Error bars denote the standard error of the mean.
While the double-imaging analysis enables to quantify the reconstruction fidelity, it is primarily sensitive to statistical errors stemming from a finite SNR. An analysis of the algorithm with simulated fluorescence images for our experimental resolution and SNR suggests that we do not expect large systematic errors (see Supplementary Note 3 for details). However, there is currently no straightforward method to accurately quantify systematic errors with experimental data.
Conclusions
We have presented an unsupervised deep-learning algorithm for the reconstruction of the lattice occupation from fluorescence images obtained with quantum gas microscopes. Using experimental images from our cesium experiment, we demonstrated high-fidelity reconstruction of the site occupation using the proposed algorithm in a challenging regime, where the lattice spacing is more than two times smaller than the imaging resolution. Based on a convolutional neural network that is able to efficiently approximate a wide variety of nonlinear relationships, our approach has an inherent advantage in solving the deconvolution problem and capturing density-dependent effects12,26. The autoencoder architecture allows training directly with experimental images, using a dataset with about 200 fluorescence images that can be taken within a few hours in a typical microscope setup. This also obviates the need for a detailed calibration of imaging parameters such as PSF or image noise. The well-optimized tensor operations allow the reconstruction of a full image in less than a second on standard computer hardware. Finally, we have developed and implemented methods for benchmarking the reconstruction performance, providing a path to estimate the fidelity with experimental data where the true occupation is a priori not known. In particular, we have shown how a network structure as employed here allows one to extract the bimodal distribution of deconvolved counts before the final binarization step. This carries a lot of information about the reconstruction process, facilitating an evaluation of the reconstruction fidelity as well as the detection of possible drifts in a day-to-day operation. Additionally, we demonstrated how a double-imaging protocol can be used to estimate the reconstruction fidelity in the presence of finite hopping and losses during imaging.
Our work enables high-fidelity reconstruction in experiments with short-spaced optical lattices, paving the way for new experiments with exotic lattice configurations34,35,36. We anticipate our scheme to remain applicable for even shorter lattice spacings, as long as the SNR is increased appropriately. Beyond the field of quantum gas microscopy, this algorithm can also be applied to other experimental platforms, such as Rydberg-atom arrays53,54 or ion trap experiments55.
Methods
Experimental setup
The science chamber is a glass cell equipped with an external high-resolution objective at a numerical aperture of 0.8. The atoms are loaded into a 2D square optical lattice created from the interference of two perpendicular retro-reflected laser beams with wavelength λ = 767 nm (spacing a = 383.5 nm). We perform fluorescence imaging by suddenly increasing the lattice depth to about 400 μK and adding optical molasses on the D2 line (λ = 852 nm). The scattered fluorescence photons are collected through the microscope objective and focused onto an sCMOS camera. With this imaging system, a resolution of about 850 nm is obtained.
To prepare experimental images at different filling fractions, we start from unity-filling Mott insulators, and then randomly remove a certain fraction of atoms by driving coherent microwave transfers on the F = 3, mF = 3 ↔ F = 4, mF = 4 transition for a variable duration and then removing the transferred atoms using an optical blowout pulse (resonant with the \(F=4\leftrightarrow {F}^{{\prime} }=5\) transition).
Lattice extraction
The reconstruction scheme relies on the fact that the lattice sites are always at the same position in each processed crop. To determine the lattice grid, we first extract the lattice base vectors from a set of dilute fluorescence images. In each image, isolated atoms are fitted with 2D Gaussians to determine their location with sub-pixel accuracy. Fourier transforming the fitted positions and averaging over all images yields distinct peaks, corresponding to the lattice wave vectors. Finally, the phase in each analyzed image is fixed by fitting a grid spanned by the previously determined base vectors to the positions of a few isolated atoms around the central region of interest.
Data processing
For training and reconstruction, experimental images are first rotated to align the lattice axes close to horizontal/vertical, and then cut into crops containing exactly 16 × 16 lattice sites. Furthermore, the pixel values are re-scaled to the range (−1,1) using a linear minimum-maximum scaling, where the minimum and maximum values are determined across the entire training set. The same scaling is applied to each processed crop.
Data availability
The data that supports the plots within this paper and other findings of this study are available at https://doi.org/10.17617/3.RNICHV.
Code availability
The code for the model and evaluation within this paper are available from the corresponding author upon reasonable request.
References
Carleo, G. et al. Machine learning and the physical sciences. Rev. Mod. Phys. 91, 045002 (2019).
Carrasquilla, J. Machine learning for quantum matter. Adv. Phys. X 5, 1797528 (2020).
Rem, B. S. et al. Identifying quantum phase transitions using artificial neural networks on experimental data. Nat. Phys. 15, 917–920 (2019).
Käming, N. et al. Unsupervised machine learning of topological phase transitions from experimental data. Mach. Learn. Sci. Technol. 2, 035037 (2021).
Bohrdt, A. et al. Classifying snapshots of the doped Hubbard model with machine learning. Nat. Phys. 15, 921–924 (2019).
Khatami, E. et al. Visualizing strange metallic correlations in the two-dimensional fermi-hubbard model with artificial intelligence. Phys. Rev. A 102, 033326 (2020).
Wigley, P. B. et al. Fast machine-learning online optimization of ultra-cold-atom experiments. Sci. Rep. 6, 25890 (2016).
Tranter, A. D. et al. Multiparameter optimisation of a magneto-optical trap using deep learning. Nat. Commun. 9, 4360 (2018).
Picard, L. R. B., Mark, M. J., Ferlaino, F. & Bijnen, R. V. Deep learning-assisted classification of site-resolved quantum gas microscope images. Meas. Sci. Technol. 31, 025201 (2019).
Gross, C. & Bakr, W. S. Quantum gas microscopy for single atom and spin detection. Nat. Phys. 17, 1316–1323 (2021).
Bakr, W. S., Gillen, J. I., Peng, A., Fölling, S. & Greiner, M. A quantum gas microscope for detecting single atoms in a hubbard-regime optical lattice. Nature 462, 74–77 (2009).
Sherson, J. F. et al. Single-atom-resolved fluorescence imaging of an atomic mott insulator. Nature 467, 68–72 (2010).
Endres, M. et al. Single-site- and single-atom-resolved measurement of correlation functions. Appl. Phys. B 113, 27–39 (2013).
Cheuk, L. W. et al. Quantum-gas microscope for fermionic atoms. Phys. Rev. Lett. 114, 193001 (2015).
Edge, G. J. A. et al. Imaging and addressing of individual fermionic atoms in an optical lattice. Phys. Rev. A 92, 063406 (2015).
Haller, E. et al. Single-atom imaging of fermions in a quantum-gas microscope. Nat. Phys. 11, 738–742 (2015).
Omran, A. et al. Microscopic observation of pauli blocking in degenerate fermionic lattice gases. Phys. Rev. Lett. 115, 263001 (2015).
Parsons, M. F. et al. Site-resolved imaging of fermionic 6-Li in an optical lattice. Phys. Rev. Lett. 114, 213002 (2015).
Yamamoto, R., Kobayashi, J., Kuno, T., Kato, K. & Takahashi, Y. An ytterbium quantum gas microscope with narrow-line laser cooling. N. J. Phys. 18, 023016 (2016).
Mitra, D. et al. Quantum gas microscopy of an attractive Fermi-Hubbard system. Nat. Phys. 14, 173–177 (2018).
Kwon, K., Kim, K., Hur, J., Huh, S. & Choi, J.-y. Site-resolved imaging of a bosonic Mott insulator of 7-Li atoms. Phys. Rev. A 105, 033323 (2022).
Yang, J., Liu, L., Mongkolkiattichai, J. & Schauss, P. Site-resolved imaging of ultracold fermions in a triangular-lattice quantum gas microscope. PRX Quantum 2, 020344 (2021).
Li, M.-D. et al. High-powered optical superlattice with robust phase stability for quantum gas microscopy. Opt. Express 29, 13876–13886 (2021).
La Rooij, A., Ulm, C., Haller, E. & Kuhr, S. A comparative study of deconvolution techniques for quantum-gas microscope images. Preprint at arXiv:2207.08663 (2022).
Greif, D. et al. Site-resolved imaging of a fermionic mott insulator. Science 351, 953–957 (2011).
Masson, S. J. & Asenjo-Garcia, A. Universality of Dicke superradiance in arrays of quantum emitters. Nat. Commun. 13, 2285 (2022).
Wiener, N. Extrapolation, Interpolation, and Smoothing of Stationary Time Series (MIT Press, 1964).
Richardson, W. H. Bayesian-based iterative method of image restoration. JOSA 62, 55–59 (1972).
Lucy, L. B. An iterative technique for the rectification of observed distributions. Astron. J. 79, 745 (1974).
Alberti, A. et al. Super-resolution microscopy of single atoms in optical lattices. N. J. Phys. 18, 053010 (2016).
Starck, J. L., Pantin, E. & Murtagh, F. Deconvolution in astronomy: a review. Publ. Astron. Soc. Pac. 114, 1051 (2002).
Arridge, S., Maass, P., Öktem, O. & Schönlieb, C. B. Solving inverse problems using data-driven models. Acta Numer. 28, 1–174 (2019).
Genzel, M., Macdonald, J. & März, M. Solving inverse problems with deep neural networks – Robustness included? IEEE Trans. Pattern Anal. Mach. Intell. 45, 1119–1134 (2023).
Yamamoto, R., Ozawa, H., Nak, D. C., Nakamura, I. & Fukuhara, T. Single-site-resolved imaging of ultracold atoms in a triangular optical lattice. N. J. Phys. 22, 123028 (2020).
Garwood, D., Mongkolkiattichai, J., Liu, L., Yang, J. & Schauss, P. Site-resolved observables in the doped spin-imbalanced triangular Hubbard model. Phys. Rev. A 106, 013310 (2022).
Mongkolkiattichai, J., Liu, L., Garwood, D., Yang, J. & Schauss, P. Quantum gas microscopy of a geometrically frustrated Hubbard system. Preprint at arXiv:2210.14895 (2022).
Robens, C. et al. Low-entropy states of neutral atoms in polarization-synthesized optical lattices. Phys. Rev. Lett. 118, 065302 (2017).
Baier, S. et al. Realization of a strongly interacting Fermi gas of dipolar atoms. Phys. Rev. Lett. 121, 093602 (2018).
Chomaz, L. et al. Dipolar physics: a review of experiments with magnetic quantum gases. Rep. Prog. Phys. 86, 026401 (2022).
Van Der Maaten, L., Postma, E. & Van den Herik, J. Dimensionality reduction: a comparative review. J. Mach. Learn. Res. 10, 13 (2009).
Masci, J., Meier, U., Cireşan, D. & Schmidhuber, J. Stacked convolutional auto-encoders for hierarchical feature extraction. In ICANN 2011 (eds Honkela, T., Duch, W., Girolami, M. & Kaski, S.) 52–59 (Springer, 2011).
Bengio, Y., Courville, A. & Vincent, P. Representation learning: a review and new perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 35, 1798–1828 (2013).
Nguyen, Q. & Hein, M. Optimization landscape and expressivity of deep CNNs. In Proc. 35th International Conference on Machine Learning 3730–3739 (PMLR, 2018).
Bianchini, M. & Scarselli, F. On the complexity of neural network classifiers: a comparison between shallow and deep architectures. IEEE Trans. Neural Netw. Learn. Syst. 25, 1553–1565 (2014).
Simonyan, K. & Zisserman, A. Very deep convolutional networks for large-scale image recognition. In 3rd International Conference on Learning Representations (ICLR2015), 1–14 (2015).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. ImageNet classification with deep convolutional neural networks. Commun. ACM 60, 84–90 (2017).
Springenberg, J. T., Dosovitskiy, A., Brox, T. & Riedmiller, M. Striving for simplicity: the all convolutional net. In 3rd International Conference on Learning Representations (ICLR2015), 1–14 (2015).
Salakhutdinov, R. & Hinton, G. Semantic hashing. Int. J. Approx. Reason. 50, 969–978 (2009).
Klostermann, T. et al. Fast long-distance transport of cold cesium atoms. Phys. Rev. A 105, 043319 (2022).
Klostermann, T. M. Construction of a Caesium Quantum Gas Microscope. Ph.D. thesis, Ludwig-Maximilians-Universität (2022).
von Raven, H. A New Caesium Quantum Gas Microscope with Precise Magnetic Field Control. Ph.D. thesis, Ludwig-Maximilians-Universität (2022).
Kingma, D. P. & Ba, J. Adam: a method for stochastic optimization. In 3rd International Conference on Learning Representations (ICLR2015), 1–13 (2015).
Saffman, M., Walker, T. G. & Mølmer, K. Quantum information with Rydberg atoms. Rev. Mod. Phys. 82, 2313–2363 (2010).
Morgado, M. & Whitlock, S. Quantum simulation and computing with Rydberg-interacting qubits. AVS Quantum Sci. 3, 023501 (2021).
Bruzewicz, C. D., Chiaverini, J., McConnell, R. & Sage, J. M. Trapped-ion quantum computing: Progress and challenges. Appl. Phys. Rev. 6, 021314 (2019).
Acknowledgements
The authors would like to thank Christian Schweizer for their contributions to the early stages of this experiment and Annabelle Bohrdt for helpful discussions on machine learning architectures. A.I. and T.K. were supported by the Bavarian excellence network ENB via the International Ph.D. Program of Excellence Exploring Quantum Matter (ExQM). J.F.W. acknowledges support from the German Academic Scholarship Foundation and the Marianne-Plehn-Program. H.v.R. acknowledges support from the Hector Fellow Academy. C.R.C. has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No 897142 and the MCQST seed funding (EXC-2111/Projekt-ID: 390814868). We acknowledge funding from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) via Research Unit FOR 2414 under project number 277974659 and under Germany’s Excellence Strategy - EXC-2111 - 390814868 and from the German Federal Ministry of Education and Research via the funding program quantum technologies—from basic research to market (contract number 13N15895 FermiQP).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
All authors contributed substantially to the work presented in this manuscript. A.I. and S. Hae. devised the reconstruction method and performed it together with J.F.W. the data analysis. A.I. and J.F.W. carried out the experimental measurements. A.I., J.F.W., S.Hae., H.v.R., S.Hu., T.K., C.R.C., M.A., and I.B. contributed to the construction and design of the experimental setup. I.B. and M.A. supervised the study. All authors contributed to the writing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Physics thanks Christian Roos and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Impertro, A., Wienand, J.F., Häfele, S. et al. An unsupervised deep learning algorithm for single-site reconstruction in quantum gas microscopes. Commun Phys 6, 166 (2023). https://doi.org/10.1038/s42005-023-01287-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42005-023-01287-w
This article is cited by
-
An atomic boson sampler
Nature (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.