Abstract
Adhesion-regulating molecule 1 (ADRM1) has been implicated in tumor development, yet its specific role in bladder cancer (BC) remains undefined. This study aimed to elucidate the function of ADRM1 in BC through a combination of bioinformatics analysis and immunohistochemical analysis (IHC). Utilizing R version 3.6.3 and relevant packages, we analyzed online database data. Validation was conducted through IHC data, approved by the Institutional Ethics Committee (Approval No. K20220830). In both paired and unpaired comparisons, ADRM1 expression was significantly elevated in BC tissues compared to adjacent tissues, as evidenced by the results of TCGA dataset and IHC data. Patients with high ADRM1 expression had statistically worse overall survival than those with low ADRM1 expression in TCGA dataset, GSE32548 dataset, GSE32894 dataset, and IHC data. Functional analysis unveiled enrichment in immune-related pathways, and a robust positive correlation emerged between ADRM1 expression and pivotal immune checkpoints, including CD274, PDCD1, and PDCD1LG2. In tumor microenvironment, samples with the high ADRM1 expression contained statistical higher proportion of CD8 + T cells and Macrophage infiltration. Meanwhile, these high ADRM1-expressing samples displayed elevated tumor mutation burden scores and stemness indices, implying potential benefits from immunotherapy. Patients with low ADRM1 expression were sensitive to cisplatin, docetaxel, vinblastine, mitomycin C, and methotrexate. According to the findings from bioinformatics and IHC analyses, ADRM1 demonstrates prognostic significance for BC patients and holds predictive potential for both immunotherapy and chemotherapy responses. This underscores its role as a biomarker and therapeutic target in BC.
Similar content being viewed by others
Introduction
Bladder cancer (BC) stands as one of the most common malignancies affecting the urinary system1,2. In 2022 approximately 81,180 new cancer cases and 17,100 cancer deaths in the United States2,3. Of these, BC can clinically be divided into two subtypes: muscle-invasive bladder cancer (MIBC) and non-muscle-invasive bladder cancer (NMIBC), based on whether tumors invade the detrusor muscle4,5. Despite the continuous development of treatments, such as surgical interventions, Bacillus Calmette-Guerin (BCG), chemotherapy, and immunotherapy, the prognosis and treatment methods of BC are still far from satisfactory6,7. For instance, radical surgery (RC) is the standard treatment for MIBC. However, this therapy would bring heavy economic, physical, and mental burdens to patients8,9. Although experiencing a traumatic therapy, MIBC patients will recur rapidly following RC9,10. In addition to surgery, many patients are resistant to drugs (such as BCG, chemotherapy), which predict a poor prognosis of BC patients11,12. Meanwhile, some of the respondents to these treatments had to compelled to discontinue due to severe treatment-related adverse events13. Thus, enhancing the prognosis of BC patients has prompted researchers to explore new therapeutical targets and optimize the utilization of existing therapies through the identification of robust biomarkers14.
Adhesion-regulating molecule 1 (ADRM1), a ubiquitin receptor located on the 26S proteasome15, play a role in the cell adhesion, deubiquitination, and proteolysis16. Within the context of human cancers, deubiquitinating proteins are increasingly recognized as pivotal oncogenes or tumor suppressor genes across various malignancies17. Notably, recent studies have highlighted a correlation between elevated ADRM1 expression and unfavorable overall survival (OS) in distinct malignancies, such as hepatocellular carcinoma18, gastric cancer19, and breast cancer20. Specifically, inhibiting ADRM1 would suppress cancer cells growth in vitro18,21. Furthermore, ADRM1 also encodes RPN13, a critical ubiquitin receptor, and the application of inhibitors targeting the ADRM1/RPN13 interaction has been shown exert a substantial restraining effect on the amplification of ovarian cancer cell22. Remarkably, these inhibitors have displayed the capacity to induce a synergistic cytotoxic response in ovarian cancer cell in conjunction with cisplatin or doxorubicin22. In the tumor microenvironment of ovarian cancer, RPN13/ADRM1 inhibitors can reverse immunosuppression which effects attributed to myeloid-derived suppressor cells23. All these results underscore the potential of ADRM1 as a promising prognostic biomarker and a potential new therapeutic target. However, the role of ADRM1 in the context of BC remains relatively unexplored in the current literature.
Therefore, in this study, we aim to evaluate the correlation between ADRM1 and BC through a comprehensive approach involving bioinformatics analysis and immunohistochemical analysis (IHC). Specifically, our study pursues two primary objectives: firstly, to scrutinize the expression profile of ADRM1 in BC and its prognostic implications; secondly, to explore potential associations between ADRM1 expression, the immune microenvironment, and chemotherapy responsiveness. For validation the results of bioinformation analysis, we performed IHC using the BC samples in our institution, with ethical clearance from the appropriate committee.
Materials and methods
Online data acquisition
The Cancer Genome Atlas (www.gdc.cancer.gov, TCGA)24 repository provided both the expression and clinical data encompassing BC tissues and adjacent non-tumor tissues. Then, the ‘limma’ package was employed to compare ADRM1 expression between 19 adjacent non-tumor samples and 414 BC samples, as previous description26. At same time, an investigation into ADRM1 expression in pan-cancer was undertaken. As external validations, GSE32548 and GSE32894 datasets were extracted from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov, GEO)27. To evaluated the prognostic value of ADRM1, Kaplan‒Meier curves was use to compare the survival outcome among the high and low ADRM1 expression groups within the TCGA, GSE32548, and GSE32894 datasets. Moreover, we extended this prognostic evaluation to subgroups within these datasets.
Functional analysis
We conducted Gene Ontology (GO) enrichment analysis and the Kyoto Encyclopedia of Genes and Genomes (KEGG)28,29,30 enrichment analysis were performed to explore the potential function of ADRM1 in BC. The outcomes of these analyses, characterized by P values < 0.05 and Q values < 0.05, were presented through bubble plots. The ADRM1-related pathways were enriched using Gene Set Enrichment Analysis (GSEA) based on the REACTOME pathways. Pathways with P values < 0.05 and false discovery rate (FDR) < 25% were displayed. Meanwhile, GeneMANIA (www.genemania.org)31 was employed to screen the interacting proteins of ADRM1.
Immune-related analysis and chemotherapy prediction
Considering the study report23 and functional results indicate that ADRM1 regulates immune environment, we explore the role of ADRM1 in immune-related analysis. Initially, we compared the expression of various immune checkpoints within high and low ADRM1 expression groups. We compared the expression profiles of immune cell marker genes, extracted from CellMarker 2.032 and the study by Zhang et al.33, between the high and low ADRM1 expression groups. Then, a comparative analysis of the composition of infiltrated immune cells were compared between the low and high ADRM1 expression groups using the CIBERSORT algorithm. In order to corroborate the findings from CIBERSORT, the TIP (Tracking Tumor Immunophenotype) methodology was employed to evaluate the presence of infiltrating immune cells within each sample34. Furthermore, the tumor mutational burden (TMB) for each BC sample were calculated using the “maftools” package, based on data downloaded from TCGA database. The TMB scores of the low and high ADRM1 expression groups were subject to comparison. To gauge cellular stemness, an mRNA expression‐based stemness index (mRNAsi) was established through a one-class logistic regression machine learning algorithm35. Cellular stemness has been recognized to play a significant role in influencing the response to chemotherapy and immunotherapy35,36. Thus, we assessed the relationship between the mRNAsi score and ADRM1 expression37,38. A comparison of response rates between the high and low ADRM1 expression groups was conducted using data from the IMvigor210 trial (EGAD00001003977), encompassing patients who had all received anti-PD-L1 agent (atezolizumab). In chemotherapy prediction, the “pRRophetic” package was used to calculate the half-maximal inhibitory concentration (IC50), a key indicator of drug sensitivity. The IC50 scores were compared between the low and high ADRM1 expression groups.
Patients and clinical specimens
Between 2017 and 2019, a total of 195 BC patients accepted surgery at our hospital were enrolled in this study. There were 195 BC samples and 34 adjacent non-tumor tissues collected from the enrolled patients. Inclusion criteria stipulated that patients were clinically diagnosed and with no other malignancy history, age less than 18 or postoperative survival period less than 30 days. Clinical features including age, sex, TNM stages and so on, as previous description39. Clinical stages were determined according to the criteria of the 8th edition of AJCC/UICC system (AJCC, American Joint Committee on Cancer /UICC, Union for International Cancer Control). Our study was approved by the Ethical Committee of our institution, and the ethics review(K20220830), and all participants provided written informed consent.
Immunohistochemical analysis
IHC was performed on paraffin-embedded 3 μm sections. Following deparaffinization with xylene and rehydration with a gradient of alcohol, antigen retrieval was carried out through heating in citrate buffer. To quench endogenous peroxidase activity, 3% H2O2 was applied, followed by blocking with 5% BSA. Subsequently, the sections were incubated with a primary antibody against ADRM1 (diluted at 1:800, #17,054–1-AP, Proteintech, China) for 1.5 h at room temperature. After three washes, the sections were covered with HRP-conjugated secondary antibody at room temperature for 30 min. Thereafter, specimens were stained with 3′,3-diaminobenzidine tetrahydrochloride, counterstained with hematoxylin. After alcohol dehydration, xylene vitrification and neutral gum seal, sections were photographed using Leica 2500 microscope.
To semi-quantitatively represent the ADRM1 immunostaining results, evaluable sections were classified into four IHC scores according to the percentage of staining positive cells within the total cancerous cells: IHC score 0, 0% positive; IHC score 1, 1–10% positive; IHC score 2, 11–50% positive; IHC score 3, > 50% positive.
Statistical analysis
According to the normality and quality of variances of the data, one-way ANOVA or the Mann‒Whitney U test was used to perform statistical analysis of three or more continuous variables. Quantitative data in two groups were compared using Student’s t test. All analyzed data are displayed as the standard deviation (SD). A p < 0.05 was considered significant for all analyses, which were performed using R version 3.6.3 and relative packages. ns, p ≥ 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Ethical considerations
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics board of Taizhou Hospital, Wenzhou Medical University (K20220830), and informed consent was obtained from all patients.
Results
The basic data of ADRM1 in BC
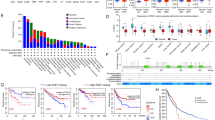
As shown in Fig. 1A, B, ADRM1 was highly expressed in many cancers than counterpart normal tissues in both unpaired and paired samples, except kidney chromophobe, pheochromocytoma, and paraganglioma. Notably, the analysis revealed a statistically significant upregulation of ADRM1 expression in BC tissues compared to normal tissues, as depicted in both unpaired and paired comparisons (Fig. 1C, D).
The expression of ADRM1 in unpaired (A) and paired (B) pan-cancer tissues. The expression of ADRM1 in unpaired (C) and paired (D) BC tissues. The diagnostic value of ADRM1 (E). The prognostic value of ADRM1 in TCGA (F), GSE32548 (G), and GSE32894 (H) datasets. ns, p ≥ 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Utilizing the ROC curve analysis, ADRM1 exhibited a moderately robust area under the curve (AUC) value of 0.87 (Fig. 1E). According to the median ADRM1 expression values extracted from the TCGA, GSE32548, and GSE32894 datasets, BC patients were divided into a high ADRM1 expression group (above the median) and a low ADRM1 expression group (below the median), respectively. As shown in Table 1, high ADRM1 expression was significantly associated with worse OS (p = 0.029) than low ADRM1 expression in the TCGA dataset. The detail information of GSE32548 and GSE32894 datasets were displayed in Supplementary Tables 1 and 2, respectively.
ADRM1predictes the prognosis of BC patients
The Kaplan–Meier curves clearly indicate a significant association between high ADRM1 expression and poor OS in BC patients (Fig. 1F, p = 0.014) in the TCGA dataset. In external validations, patients with high ADRM1 expression also had shorter survival time than those with low ADRM1 expression in the GSE32548 (Fig. 1G, p = 0.007) and GSE32894 (Fig. 1H, p = 0.023) cohorts. Subgroup analyses also underscore this correlation, revealing high ADRM1 expression as significantly linked to adverse OS outcomes in several subgroups: World Health Organization (WHO) high grade (Fig. 2A, p = 0.01), T3-4 stage (Fig. 2B, p = 0.01), N1-3 stage (Fig. 2C, p = 0.036), pathological stage III-IV (Fig. 2D, p = 0.004), male patients (Fig. 2E, p = 0.05), and patients below 70 years old (Fig. 2F, p = 0.033).
The prognostic value of ADRM1: TCGA subgroups [WHO high grade (A), T3_4 stage (B), lymph node metastasis (C), pathological stage (D), Male (E), and age < = 70 years (F)]; GSE32584 subgroups [WHO low grade (G), male (H), and age < = 70 years (I)]. GSE32894 subgroups [WHO G3 (J), T1_4 (K), and age < = 70 years (L)].
In the external GSE32548 dataset, compared with the low ADRM1 expression group, the high ADRM1 expression group was significantly correlated with shorter OS in subgroups: WHO low grade (Fig. 2G, p = 0.039), male patients (Fig. 2H, p = 0.019) and patients below 70 years old (Fig. 2I, p = 0.035). In external GSE32984 dataset, ADRM1 also presented significant prognostic value in subgroups, such as WHO G3 (Fig. 2J, p = 0.036), T1_4 (Fig. 2K, p = 0.006) and patients below 70 years old (Fig. 2L, p = 0.011). These results revealed that ADRM1 expression might be a prognostic factor for BC patients.
ADRM1 involves in immune-related pathways
To further explore the potential function of ADRM1 in BC, a series of functional analyses performed utilizing ADRM1 differential genes based on the TCGA dataset. The GO enrichment analysis highlighted ADRM1's significant involvement in hormone metabolic process, apical part of cell, apical plasma membrane, receptor ligand activity and others (Fig. 3A). The KEGG pathway analysis underscored ADRM1's enrichment in pathways such as chemical carcinogenesis, metabolism of xenobiotics by cytochrome P450, and so on (Fig. 3B).
Subsequently, a GSEA analysis was conducted to discern the REACTOME pathways that exhibited differential regulation based on ADRM1 differentially expressed genes within the high and low ADRM1 expression groups. Figure 3C showed that ADRM1 differentially expressed genes were found to enriched immune-related categories, such as interleukin 1 signaling, interleukin 1 family signaling, PD1 signaling, and interleukin 12 family signaling pathways. Meanwhile, the GSEA revealed enrichments in processes such as biological oxidations, glucuronidation, miscellaneous substrates and nuclear receptor transcription pathways (Fig. 3D). According to the GeneMANIA results, we found that ADRM1 protein was associated with specific proteins such as UBB, UBC, NOS2, and PSMA8 (Fig. 3E).
The immunological role of ADRM1 and ADRM1 predicts the response of chemotherapeutic drugs
The GSEA analysis prominently highlighted numerous gene sets enriched immune-related pathways, prompting a deeper investigation into the relationship between ADRM1 and immunological processes within the context of BC, utilizing data from the TCGA database. In high ADRM1 expression group, the expression of some important immune checkpoints increased, such as CD274 (PD-L1), CTLA4, PDCD1 (PD-1) and CDCD1LG2 (PD-L2) (Fig. 4A). The expression of cell markers notably demonstrated a substantial association between heightened ADRM1 expression and increased T cell infiltration within the high ADRM1 expression group (Fig. 4B). As the results unveiled a markedly elevated proportion of CD8 + T cells and M1 Macrophage in the high ADRM1 expression group, whereas a comparatively higher abundance of B plasma cell, CD4 + T memory resting cells and activated Mast cells was presented in the low ADRM1 expression group (Fig. 4C). In a parallel vein, the TIP analysis corroborated these findings, highlighting a significantly augmented proportion of CD8 + T naïve cells and CD4 + memory cells within the high ADRM1 expression group (Fig. 4D).
Immune-related analysis based on TCGA dataset: immune checkpoints (A), cell marker (B), immune infiltration on CIBERSORT (C) and TIP (D), the tumor mutation burden between high and low ADRM1 expression groups (E), the stemness index between high and low ADRM1 expression groups (F). The immunotherapy response in IMvigor210 (G). The IC50 of chemotherapy between the low and high ADRM1 groups (H). IC50: half-maximal inhibitory concentration.
According to the low and high ADRM1 expression groups, we compared TMB score of BC. It's probably worth noting that patients in the high ADRM1 expression group had higher TMB score than those in the low ADRM1 expression group (Fig. 4E, p = 0.039). To explore mRNAsi in BC, we compared the expression of mRNAsi between the low and high ADRM1 expression groups. As shown, a significant difference in stemness indices within the low and high ADRM1 expression group, which suggested that the high ADRM1 expression group exhibited higher stemness than the low ADRM1 expression group (Fig. 4F, p < 0.001).
Patients with response had higher mean ADRM1 expression, whereas it did not reach a significant result (Fig. 4G). As the results illustrated that the IC50 values of cisplatin, docetaxel, vinblastine, mitomycin C and methotrexate in the high ADRM1 expression group were lower than that in the low ADRM1 expression group (Fig. 4H). These suggested that high expression of ADRM1 might poorly respond to chemotherapy.
ADRM1 is a biomarker in real-world data
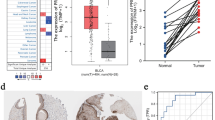
Considering the previously observed elevation of ADRM1 mRNA in breast cancer (BC) patients, we extended our investigation to evaluate ADRM1 protein expression. Immunohistochemistry was conducted on tumor tissues from BC patients, and ADRM1 protein levels were assessed and scored in both tumor and adjacent non-tumor tissues. ADRM1 was predominantly localized within the nuclei of cancer cells. In the non-neoplastic surrounding urothelium, immunostaining for ADRM1 protein was mostly absent or exhibited weak intensity in most instances. (Fig. 5A). Immunostaining revealed higher protein expression for ADRM1 in most of the tumor tissues of BC patients in both unpaired (Fig. 5B) and paired (Fig. 5C) comparisons.
Validation the bioinformatics results of ADRM1: representative IHC staining for ADRM1 in BC and adjacent non-tumor tissues (A); the expression of ADRM1 in unpaired (B) and paired (C) tissues; the prognostic value of ADRM1 in IHC data: overall survival (D), subgroups [WHO high grade (E), Ta_1 stage (F), no lymph node metastasis (G), no distant metastasis (H), male (I), and age < = 70 years (J)]; WHO: World Health Organization, IHC: Immunohistochemical analysis. ns, p ≥ 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Based on the immunostaining intensity for ADRM1 in the tumor tissues, patients were categorized into low ADRM1 group (including patients scored by 0–2) and high ADRM1 group (including patients scored by 3). As shown in Table 2, the expression level of ADRM1 protein was observed to be associated with age, WHO grade, T stage and OS (Table 2).
As shown in Fig. 5D, Kaplan–Meier survival curves demonstrated that the high ADRM1 group had a shorter survival time than the low ADRM1 group (p < 0.001). To further examine the correlation between the expression of ADRM1 protein level and prognosis, Kaplan–Meier survival curves was employed to evaluated the prognostic value of ADRM1 in subgroups. In subgroups, ADRM1 also presented significant prognostic value in WHO high grade (Fig. 5E, p < 0.001), Ta_1 (Fig. 5F, p = 0.035), no lymph node metastasis (Fig. 5G, p < 0.001), no distant metastasis (Fig. 5H, p < 0.001), male (Fig. 5I, p < 0.001) and patients below 70 years old (Fig. 5J, p = 0.024). Collectively, the data from our institution corroborates the prognostic value of ADRM1 in predicting outcomes for BC patients.
Discussion
ADRM1 is a ubiquitin receptor on the 26S proteasome, which is activated by the binding of ubiquitin and S1 subunit to the 19S complex15. This complex recruits the deubiquitinating enzyme UCH37 to the 26S proteasome40. In tumors, the 12 prognosis-related genes including ADRM1 might be promising therapeutic targets esophageal adenocarcinoma41. RA190, a specific inhibitor of ADRM1, induces apoptosis intrahepatic cholangiocarcinoma cells42. Moreover, ADRM1 interference combined with 5-fluorouracil treatment efficiently suppressed colorectal cancer cell growth in vitro21. These data support that ADRM1 may be essential for the maintenance of the malignant status of cells and thus affects the prognosis and treatment effect. Hence, we systematically investigated the role of ADRM1 in BC.
In this study, we investigated the relationship between ADRM1 expression and clinicopathological parameters and patient survival outcomes based on the TCGA database and two independent GEO datasets. The results revealed that ADRM1 mRNA significantly overexpressed in many tumors, including BC. Meanwhile, we adopted IHC to validate the results, which also identified that ADRM1 was highly expressed in BC tissues. Furthermore, high ADRM1 expression was associated with worse OS in BC patients. Subgroup analysis of both TCGA group and independent GEO datasets showed that a high ADRM1 expression was significantly correlated with a poor prognosis in BC in versus clinical subgroups. In our IHC data, ADRM1 predicted prognosis in whole cohort or subgroup of BC patients. Similarly, a study based on bioinformatics research also reported that a group of genes including ADRM1, PPARD, CST4, CSNK1E, PTPN14 and ETV6 could be a potential biomarker group for BC43. These results supported the idea that ADRM1 indeed be involved in the pathogenesis of BC, and it is more likely to be highly expressed of ADRM1 often predicts a poor prognosis.
We then applied function analysis, GO, KEGG, and GSEA, to explore the possible functions and mechanisms of action of ADRM1. Immune-related pathways were highly associated with the high ADRM1 expression group. According to the GeneMANIA results, we found that ADRM1 proteins were associated with proteins like UBB, UBC, NOS2, PSMA8 and PSMD2. UBB and UBC are two ubiquitin gene, which code for poly-Ub precursors. UBB or UBC protein was required for some cancer cells to keep tumor features, like myeloma cells44. NOS2 is an adaptive immune pathway gene, together with CD8A, CD68 and GZMB, show significant positive correlation with most of immune checkpoint coding genes in hepatocellular carcinoma45. PSMA8 is a member of PSMA family. The PSMA family genes were positively correlated with the cell cycle, ubiquinone metabolism, and immune response signaling46. PSMD2 expression was reported be correlated with immune cell infiltration in lung adenocarcinoma46. All these results shown that ADRM1 is highly likely to be an immune-related gene.
As the development of checkpoint inhibitor therapy, five immunotherapy agents targeting the primarily targeting programmed cell death-1 protein (PD-1) or its ligand (PD-L1) pathway have been approved by the US Food and Drug Administration (FDA) for BC patients, including atezolizumab, pembrolizumab, avelumab, nivolumab, and durvalumab. A study showed higher response rates with atezolizumab in patients with increased PD-L1 expression, compared to those with lower levels of PD-L1 expression46. In high ADRM1 expression group, the expression of PD-L1 and PD-1 increased, suggesting that patients with high ADRM1 expression were more likely to benefit from anti-PD-L1 therapy. The tumor microenvironment plays a vital role in immunotherapy47,48. In the aspect of infiltrating immune cells, the high ADRM1 group contained significantly higher proportion of CD8 + T cells and Macrophage (M1), and lower proportion of CD4 + T memory cells and Mast cells. There has reported that macrophage M1 was a predictor of immune-checkpoint blockades therapy for metastatic urothelial cancer patients25,49. Upregulated immune activation pathways observed in the high-M1 subset, which identified favorable response to immunotherapy49. Jia Lv et al. reported that BC patients with higher infiltration levels of CD8 + T cell and lower Mast cells are more likely to present with better immunotherapeutic effect and prognosis50,51,52. According to these data, we may infer that BC patients with high ADRM1 might has represent a better respond to immunotherapy than low ADRM1 patients. The high and low ADRM1 expression groups showed significant differences in TMB analysis. Higher ratio of relevant genomic alterations was found in the high ADRM1 expression group. And the expression of ADRM1 showed positive correlation with TMB score, which suggested that patients with different expression level of ADRM1 might suitable for different treatment options. Similarly, the high ADRM1 expression group was significantly correlated with higher stemness index than the low ADRM1 group. Patients with high stemness index were positively response to immunotherapy35. Thus, the results of immune-related analysis suggested that BC patients with high ADRM1 might has represent a better respond to immunotherapy than low ADRM1 patients.
Recently, a study reported that inhibition of ADRM1/RPN13 could has a synergistic cytotoxic response to ovarian cancer cell with cisplatin or doxorubicin22. Chen et al. also found that ADRM1 interference combined with 5-fluorouracil treatment efficiently suppressed colorectal cancer cell growth in vitro21. Hence, we evaluated the efficiency of chemotherapy in high and low ADRM1 expression groups. Consistent with the literatures, this research found that patients in the low ADRM1 expression group were more sensitive to multiple chemotherapy drugs. Given the results of immune-related analysis, patients could choose immunotherapy or chemotherapy as optimal treatment according to the level of ADRM1. Thus, we suggest that ADRM1 may be a useful biomarker for BC patients.
Conclusion
By bioinformatics and IHC analyses, we identified that ADRM1 had prognostic value in BC patients and could predict the immunotherapy and chemotherapy responses, indicating that it is a biomarker and target of BC. Of course, many basic and animal experiments are required to further identified these results, which also is the next work of us.
Data availability
Since human data are involved, you should contact the corresponding authors to obtain relevant information.
References
Chen, W. et al. Cancer incidence and mortality in China, 2014. Chin. J. Cancer Res. 30(1), 1–12. https://doi.org/10.21147/j.issn.1000-9604.2018.01.01 (2018).
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71(3), 209–249. https://doi.org/10.3322/caac.21660 (2021).
Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 72(1), 7–33. https://doi.org/10.3322/caac.21708 (2022).
Netto, G. J. et al. The 2022 world health organization classification of tumors of the urinary system and male genital organs-part B: Prostate and urinary tract tumors. Eur. Urol. 82(5), 469–482. https://doi.org/10.1016/j.eururo.2022.07.002 (2022).
Villoldo, G. M. et al. A Th2-score in the tumor microenvironment as a predictive biomarker of response to Bacillus Calmette Guérin in patients with non-muscle invasive bladder carcinoma: A retrospective study. Oncol Res. 31(2), 207–220. https://doi.org/10.32604/or.2023.028163 (2023).
Shih, K. W. et al. Non-muscular invasive bladder cancer: Re-envisioning therapeutic journey from traditional to regenerative interventions. Aging Dis. 12(3), 868–885. https://doi.org/10.14336/AD.2020.1109 (2021).
Li, D. X., Feng, D. C. & Deng, S. Re: Gyrithe Lynghøj Pedersen, Marie Schmidt Erikson, Karin Mogensen, Susanne Rosthøj, Gregers Gautier Hermann. Outpatient photodynamic diagnosis-guided laser destruction of bladder tumors is as good as conventional inpatient photodynamic diagnosis-guided transurethral tumor resection in patients with recurrent intermediate-risk low-grade ta bladder tumors. A prospective randomized noninferiority clinical trial. Eur Urol 2023;83:125-30. Eur. Urol. https://doi.org/10.1016/j.eururo.2023.06.029 (2023).
Li, D. X. et al. Lymphocyte-to-monocyte ratio (LMR) during induction is a better predictor than preoperative LMR in patients receiving intravesical Bacillus calmette -guerin for non-muscle-invasive bladder cancer. Front Oncol. 12, 937638. https://doi.org/10.3389/fonc.2022.937638 (2022).
Feng, D. et al. Comparative effectiveness of open, laparoscopic and robot-assisted radical cystectomy for bladder cancer: a systematic review and network meta-analysis. Minerva Urol. Nefrol. 72(3), 251–264. https://doi.org/10.23736/S0393-2249.20.03680-2 (2020).
Feng, D., Li, X., Liu, S., Han, P. & Wei, W. A comparison between limited bowel preparation and comprehensive bowel preparation in radical cystectomy with ileal urinary diversion: a systematic review and meta-analysis of randomized controlled trials. Int. Urol. Nephrol. 52(11), 2005–2014. https://doi.org/10.1007/s11255-020-02516-9 (2020).
Li, D. X., Wang, X. M., Feng, D. C. & Han, P. Neutrophil-to-lymphocyte ratio (NLR) during induction is a better predictor than preoperative NLR in non-muscle-invasive bladder cancer receiving Bacillus Calmette-GuÉRin. Asian J.Surg. 46(3), 1348–1351. https://doi.org/10.1016/j.asjsur.2022.08.108 (2023).
Zhao, M., He, X. L. & Teng, X. D. Understanding the molecular pathogenesis and prognostics of bladder cancer: an overview. Chin J Cancer Res. 28(1), 92–98. https://doi.org/10.3978/j.issn.1000-9604.2016.02.05 (2016).
EAU Guidelines. Edn. presented at the EAU Annual Congress Milan (2023).
Feng, D., Li, D., Wu, R. & Han, P. Scientific advancements in drug development and trials for urothelial Carcinoma: Insights from the 2023 ASCOGU cancers symposium. Aging Dis https://doi.org/10.14336/AD.2023.0502 (2023).
Besche, H. C. et al. Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates. EMBO J. 33(10), 1159–1176. https://doi.org/10.1002/embj.201386906 (2014).
Cherix, N. et al. A Phg2-Adrm1 pathway participates in the nutrient-controlled developmental response in Dictyostelium. Mol. Biol. Cell. 17(12), 4982–4987. https://doi.org/10.1091/mbc.e06-07-0619 (2006).
Liao, Y. et al. Deubiquitinating enzyme OTUB1 in immunity and cancer: Good player or bad actor?. Cancer Lett. 526, 248–258. https://doi.org/10.1016/j.canlet.2021.12.002 (2022).
Liang, Y. C. et al. ADRM1 as a therapeutic target in hepatocellular carcinoma. Kaohsiung J. Med. Sci. 37(1), 47–54. https://doi.org/10.1002/kjm2.12298 (2021).
Wang, J. et al. The CCDC43-ADRM1 axis regulated by YY1, promotes proliferation and metastasis of gastric cancer. Cancer Lett. 482, 90–101. https://doi.org/10.1016/j.canlet.2020.03.026 (2020).
Wu, W. et al. Prognostic and therapeutic significance of adhesion-regulating molecule 1 in estrogen receptor-positive breast cancer. Clin. Breast Cancer. 20(2), 131-144.e3. https://doi.org/10.1016/j.clbc.2019.07.009 (2020).
Chen, W., Hu, X. T., Shi, Q. L., Zhang, F. B. & He, C. Silencing of Adrm1 by RNA interference suppresses proliferation of colorectal cancer cells. Zhonghua Zhong Liu Za Zhi 31(11), 815–819 (2009) (Chinese PMID: 20137344).
Anchoori, R. K. et al. Chirality and asymmetry increase the potency of candidate ADRM1/RPN13 inhibitors. PLoS ONE 16(9), e0256937. https://doi.org/10.1371/journal.pone.0256937 (2021).
Soong, R. S. et al. RPN13/ADRM1 inhibitor reverses immunosuppression by myeloid-derived suppressor cells. Oncotarget 7(42), 68489–68502. https://doi.org/10.18632/oncotarget.12095 (2016).
Wei, L. et al. TCGA-assembler 2: Software pipeline for retrieval and processing of TCGA/CPTAC data. Bioinformatics 34(9), 1615–1617. https://doi.org/10.1093/bioinformatics/btx812 (2018).
Li, D. X. et al. M7G-related molecular subtypes can predict the prognosis and correlate with immunotherapy and chemotherapy responses in bladder cancer patients. Eur. J. Med. Res. 28(1), 55. https://doi.org/10.1186/s40001-023-01012-x (2023).
Yu, Q. et al. An inflammation-related signature could predict the prognosis of patients with kidney renal clear cell carcinoma. Front. Genet. 13, 866696. https://doi.org/10.3389/fgene.2022.866696 (2022).
Edgar, R., Domrachev, M. & Lash, A. E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucl. Acids Res. 30(1), 207–210. https://doi.org/10.1093/nar/30.1.207 (2002).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucl. Acids Res. 28(1), 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28(11), 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucl. Acids Res. 51(D1), D587–D592. https://doi.org/10.1093/nar/gkac963 (2023).
Warde-Farley, D. et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucl. Acids Res. 38(Web Server issue), W214–W220. https://doi.org/10.1093/nar/gkq537 (2010).
Hu, C. et al. Cell Marker 2.0: An updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucl. Acids Res. 51(D1), D870–D876. https://doi.org/10.1093/nar/gkac947 (2023).
Zhang, C. et al. Prioritizing exhausted T cell marker genes highlights immune subtypes in pan-cancer. iScience. 26(4), 106484. https://doi.org/10.1016/j.isci.2023.106484 (2023).
Xu, L. et al. TIP: A web server for resolving tumor immunophenotype profiling. Cancer Res. 78(23), 6575–6580. https://doi.org/10.1158/0008-5472.CAN-18-0689 (2018).
Malta, T. M. et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell 173(2), 338-354.e15. https://doi.org/10.1016/j.cell.2018.03.034 (2018).
Lee, Y. C. et al. The dynamic roles of the bladder tumour microenvironment. Nat Rev Urol. 19(9), 515–533. https://doi.org/10.1038/s41585-022-00608-y (2022).
Li, D. X. et al. A novel endothelial-related prognostic index by integrating single-cell and bulk RNA sequencing data for patients with kidney renal clear cell carcinoma. Front. Genet. 14, 1096491. https://doi.org/10.3389/fgene.2023.1096491 (2023).
Charoentong, P. et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 18(1), 248–262. https://doi.org/10.1016/j.celrep.2016.12.019 (2017).
Deng-Xiong, L. et al. Systemic immune-inflammation index (SII) during induction has higher predictive value than preoperative sii in non-muscle-invasive bladder cancer patients receiving intravesical bacillus calmette -Guerin. Clin. Genitourin. Cancer. 21(3), e145–e152. https://doi.org/10.1016/j.clgc.2022.11.013 (2023).
Yao, T. et al. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat. Cell Biol. 8(9), 994–1002. https://doi.org/10.1038/ncb1460 (2006).
Zhang, X., Yang, L., Kong, M., Ma, J. & Wei, Y. Development of a prognostic signature of patients with esophagus adenocarcinoma by using immune-related genes. BMC Bioinf. 22(1), 536. https://doi.org/10.1186/s12859-021-04456-2 (2021).
Yu, G. Y. et al. RA190, a proteasome subunit ADRM1 inhibitor, suppresses intrahepatic cholangiocarcinoma by inducing NF-KB-mediated cell apoptosis. Cell Physiol. Biochem. 47(3), 1152–1166. https://doi.org/10.1159/000490210 (2018).
Abedi, Z., MotieGhader, H., Hosseini, S. S., Sheikh Beig Goharrizi, M. A. & Masoudi-Nejad, A. mRNA-miRNA bipartite networks reconstruction in different tissues of bladder cancer based on gene co-expression network analysis. Sci. Rep. 12(1), 5885. https://doi.org/10.1038/s41598-022-09920-4 (2022).
Tsherniak, A. et al. Defining a cancer dependency map. Cell 170(3), 564-576.e16. https://doi.org/10.1016/j.cell.2017.06.010 (2017).
Xu, D. et al. Identification of immune subtypes and prognosis of hepatocellular carcinoma based on immune checkpoint gene expression profile. Biomed. Pharmacother. 126, 109903. https://doi.org/10.1016/j.biopha.2020.109903 (2020).
Chiao, C. C. et al. Prognostic and genomic analysis of proteasome 20S subunit alpha (PSMA) family members in breast cancer. Diagnostics (Basel). 11(12), 2220. https://doi.org/10.3390/diagnostics11122220 (2021).
Li, D. X. et al. Identification of endothelial-related molecular subtypes for bladder cancer patients. Front. Oncol. 13, 1101055. https://doi.org/10.3389/fonc.2023.1101055 (2023).
Xing, S., Hu, K. & Wang, Y. Tumor immune microenvironment and immunotherapy in non-small cell lung cancer: Update and new challenges. Aging Dis. 13(6), 1615–1632. https://doi.org/10.14336/AD.2022.0407 (2022).
Zeng, D. et al. Macrophage correlates with immunophenotype and predicts anti-PD-L1 response of urothelial cancer. Theranostics. 10(15), 7002–7014. https://doi.org/10.7150/thno.46176 (2020).
Escobar, G., Mangani, D. & Anderson, A. C. T cell factor 1: A master regulator of the T cell response in disease. Sci Immunol. 5(53), eabb9726. https://doi.org/10.1126/sciimmunol.abb9726 (2020).
Vahidi, Y., Faghih, Z., Talei, A. R., Doroudchi, M. & Ghaderi, A. Memory CD4+ T cell subsets in tumor draining lymph nodes of breast cancer patients: A focus on T stem cell memory cells. Cell Oncol. 41(1), 1–11. https://doi.org/10.1007/s13402-017-0352-6 (2018).
Zhang, G. et al. Clinical predictive value of naïve and memory T cells in advanced NSCLC. Front. Immunol. 13, 996348. https://doi.org/10.3389/fimmu.2022.996348 (2022).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Interpretation of the data was performed by Q.X.Y., J.C.W., H.H.Z., J.F.L., Y.Q.G. and L.X.Y.; Y.Q.G., J.F.L. and L.X.Y. planned and performed IHC experiments. The first draft of the manuscript was written by Q.X.Y. and J.C.W., and all authors commented on previous versions of the manuscript. The final version of the manuscript was written by H.H.Z. and J.C.W.; All authors had access to the data and read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yu, Qx., Wang, Jc., Liu, Jf. et al. Adhesion-regulating molecule 1 (ADRM1) can be a potential biomarker and target for bladder cancer. Sci Rep 13, 14803 (2023). https://doi.org/10.1038/s41598-023-41992-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-41992-8
This article is cited by
-
An endothelial-related prognostic index for bladder cancer patients
Discover Oncology (2024)
-
What’s new about the tumor microenvironment of urothelial carcinoma?
Clinical and Translational Oncology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.