Abstract
Extracellular contractile injection systems (eCISs) are structurally similar to headless phages and are versatile nanomachines conserved among diverse classes of bacteria. Herein, Streptomyces species, which comprise filamentous Gram-positive bacteria and are ubiquitous in soil, were shown to produce Streptomyces phage tail-like particles (SLPs) from eCIS-related genes that are widely conserved among Streptomyces species. In some Streptomyces species, these eCIS-related genes are regulated by a key regulatory gene, which is essential for Streptomyces life cycle and is involved in morphological differentiation and antibiotic production. Deletion mutants of S. lividans of the eCIS-related genes appeared phenotypically normal in terms of morphological differentiation and antibiotic production, suggesting that SLPs are involved in other aspects of Streptomyces life cycle. Using co-culture method, we found that colonies of SLP-deficient mutants of S. lividans were more severely invaded by fungi, including Saccharomyces cerevisiae and Schizosaccharomyces pombe. In addition, microscopic and transcriptional analyses demonstrated that SLP expression was elevated upon co-culture with the fungi. In contrast, co-culture with Bacillus subtilis markedly decreased SLP expression and increased antibiotic production. Our findings demonstrate that in Streptomyces, eCIS-related genes affect microbial competition, and the patterns of SLP expression can differ depending on the competitor species.
Similar content being viewed by others
Introduction
In their natural habitat, microorganisms are members of densely populated microbial communities that facilitate competitions between neighboring cells for nutrients and niche spaces1,2,3. For example, despite the limited availability of nutrients, soil is one of the most microbially diverse environments, and it is densely populated with different bacterial and fungal species1,2,3. Co-culture model systems using two or more microbial species have been commonly used to investigate competitions between different microorganisms and to identify the factors that influence competitive fitness. These model system have elucidated the functions of many factors, ranging from low-molecular weight compounds to multi-component macromolecular machines1,4,5. For example, Gram-negative bacterial type VI secretion systems (T6SSs), which are widely conserved cell envelope-spanning nanomachines, reportedly play important roles in interbacterial competitions by injecting effector proteins into target cells in a cell–cell contact-dependent manner6. T6SSs are structurally and functionally homologous to contractile phage tails, indicating the evolutionary relationships of these multi-component machines7. Besides T6SSs, another class of phage tail-like nanomachines––extracellular contractile injection systems (eCISs)––have attracted increasing interest owing to their wide distribution among diverse microbes, including Gram-negative and Gram-positive bacteria and archaea8,9. Phylogenetic analyses have indicated that eCISs are evolutionarily distant from contractile phage tails and T6SSs8,9 despite these sharing many structural and mechanical features10,11. Furthermore, it is noteworthy that the currently known eCISs mediate the interkingdom interactions wherein these nanostructures modulate the cellular processes of target eukaryotes12,13,14,15. However, although eCISs reportedly play important roles in prokaryote–eukaryote interactions, current data on the structures and functions of eCISs are limited to a few Gram-negative bacterial species.
The filamentous bacteria Streptomyces, which produces therapeutic antibiotics, is known for the presence of highly conserved eCIS-related gene clusters in 94 out of 116 of their completed genome sequences9. These Gram-positive bacteria are ubiquitously found in natural environments such as soil and are well-known for their unique life cycles. First, a spore germinates and the germ tubes grow by apical tip extension. These tubes form a vegetative mycelial network, from which aerial mycelia extend into the air and ultimately form spores. Previous genomic studies have shown that eCIS-related genes in S. coelicolor A3(2), an extensively studied model of Streptomyces, are regulated by a key regulatory gene, bldA, essential for its life cycle16,17. Although these observations suggest the biological importance of eCIS-related genes in Streptomyces, there has been no direct evidence to support this hypothesis owing to the absence of apparent phenotypic changes in deletion mutants for the eCIS-like genes18,19. In addition, the products of these eCIS-related genes in Streptomyces have remained unknown.
In this study, we have shown that Streptomyces species, including S. lividans, produce Streptomyces phage tail-like particles (SLPs) from eCIS-related genes. Colonies of S. lividans strains that cannot produce these nanostructures are more severely invaded by fungi, suggesting the protective role of producing SLPs in microbial competitions. To the best of our knowledge, this is the first report to show the biological significance of the eCIS-related genes and the macromolecular structures encoded by these genes in Gram-positive bacteria. Further, depending on the competitor microorganisms, patterns of nanostructure expression and antibiotic production in S. lividans can differ. Our findings illustrate that Streptomyces species employ diverse responses, mediated by both SLPs and secondary metabolites, to microbial competitions.
Results
Streptomyces species produce SLPs
In previous studies, proteomic analyses have shown that phage-related proteins are expressed during the growth of S. coelicolor A3(2)20,21, although there is no information regarding its biological role and macromolecular structure. The gene cluster encoding these proteins is almost completely conserved in S. lividans, a close relative of S. coelicolor A3(2) (Fig. 1A). These gene clusters include the homologs of known eCIS genes encoding essential structural proteins (Fig. 1A), suggesting that phage-related genes in S. coelicolor and S. lividans encode eCIS-like nanostructures. Given that these eCIS-related genes are widely found in other Streptomyces species (Fig. 1A), we analyzed the phylogenetic relationships of the eCIS-related genes among actinomycetes and other bacterial classes. Based on a comprehensive annotation of the eCIS genes in the database of extracellular contractile injection systems9, we constructed a phylogenetic tree of eCIS-related genes encoding tube proteins15 conserved among proteobacteria, firmicutes, and actinobacteria (Fig. 1B). The eCIS-related genes in actinomycetes form a distinct clade that is phylogenetically distant from the functionally characterized eCISs (anti-feeding prophage, Photorhabdus virulence cassettes, and metamorphosis-associated contractile structures), which is consistent with that reported in previous studies8,9. Notably, the eCIS-related gene of S. griseus falls into a subclade within the actinomycetes-specific clade, whereas those of S. lividans, S. coelicolor, and S. albus are members of another subclade (Fig. 1B). In addition, an eCIS-related gene of S. avermitilis belongs to a clade that is more closely related to the eCIS genes of proteobacteria and firmicutes, implying that a horizontal gene transfer event might have occurred8,9.
Phylogenetic analysis of eCIS-related genes in Streptomyces species. eCIS-related genes are conserved among Streptomyces. (A) Organizations of eCIS-related genes that are conserved in the model Streptomyces species. (B) A phylogenetic tree of eCIS and SLP proteins. The tree was constructed based on 72 amino acid sequences of tube proteins using the neighbor-joining method. Red and black circles indicate the clades with the bootstrap values of 100 and 85–99, respectively. The evolutionary distances were computed using the p-distance method and are in the units of the number of amino acid differences per site. Afp, anti-feeding prophage; PVC, Photorhabdus virulence cassettes; MAC, metamorphosis-associated contractile structures.
Next, we investigated whether Streptomyces species produce eCIS-like nanostructures. Considering known Gram-negative bacterial eCISs are released into the extracellular milieu via cell lysis15, we initially cultivated S. lividans and S. coelicolor in liquid culture and then ultracentrifuged the culture supernatant. However, we failed to find phage tail-like structures in the resuspended pellets by transmission electron microscopy (TEM), suggesting that these Streptomyces species do not release the putative nanostructures into the extracellular milieu under the tested conditions. Given this, we cultivated S. lividans and S. coelicolor on solid media and performed mild extraction from their mycelia using lysozyme and a detergent. As a result, we found a number of bullet-like nanostructures, which are very similar to known Gram-negative bacterial eCISs11,15, in the extracts (Fig. 2A,B). These structures were not observed in the deletion mutant of S. lividans for a gene encoding a protein (SLIV_17120, SlpS) that is homologous to Gram-negative bacterial eCIS sheath proteins (Fig. 1A and Supplementary Fig. S1). Because deletion of the sheath proteins was previously shown to abort the assembly processes of eCIS particles11, our finding demonstrates that the phage tail-like structures of the tested Streptomyces species were synthesized from the conserved eCIS-related genes. Based on these observations, we named the nanostructures “Streptomyces phage tail-like particles (SLPs).”
Streptomyces species produce eCIS-like nanostructures. eCIS-like nanostructures were found in the mycelial extracts of Streptomyces species. (A) TEM of the mycelial extracts of S. lividans, S. coelicolor, and S. albus. Scale bars, 100 nm. (B) Putative structure of the eCIS-like particle of S. lividans. This model is based on Cryo-EM structures of Gram-negative bacterial eCISs (10,11). Each color of the components corresponds to that shown in Fig. 1A.
Previous studies have reported that the expression of SLP genes in S. coelicolor is regulated by SCO4263 encoding an LuxR-type transcriptional regulator which is suggested to be a SLP gene cluster-specific regulator18,19. SCO4263 contains the UUA codon, which is very rare in the Streptomyces genome that has a high G + C content, in its 5′ region, and its expression is completely dependent on bldA17, a gene encoding Leu-tRNAUUA, which is capable of translating the UUA codon; therefore, SLP genes are regulated by bldA in S. coelicolor. The SCO4263 homolog in S. lividans, namely slpR (Fig. 1A), also contains a UUA codon in its 5′ region, strongly suggesting that bldA is a key regulator of SLP expression in S. lividans as well as S. coelicolor. In fact, SLP structures were not found in deletion mutants for slpR and bldA of S. lividans (Supplementary Fig. S1). This confirmed that bldA-dependent regulation of SLP genes is a common mechanism in S. lividans and S. coelicolor.
To examine whether other Streptomyces species produce SLPs, we cultivated three model Streptomyces species (S. albus, S. griseus, and S. avermitilis), all of which have eCIS-related genes in their genome (Fig. 1A). Their mycelia grown on solid media were subjected to mild extraction, ultracentrifugation, and TEM, as described above. As a result, we could observe a number of SLP-like nanostructures in the extract of S. albus, whereas such structures were not found in the other species (Fig. 2A). These SLP-like nanostructures were not found in the deletion mutants for XNR0535 and XNR0530, slpS and slpR homologs, respectively, in the eCIS-related gene cluster of S. albus (Fig. 1A and Supplementary Fig. S1), suggesting that these nanostructures are the products of the SLP-like genes in S. albus. In addition, bioinformatic analysis using PHASTER22 suggested the absence of intact or defective phage genes in the S. albus genome, further confirming the origin of the SLP-like nanostructures in S. albus. Importantly, the slpR homolog is present in eCIS-related gene clusters of S. albus but absent in those of S. griseus and S. avermitilis (Fig. 1A), indicating that regulation systems for the expression of latter gene clusters might be different from those of SLPs. This may, in part, explain why SLPs were not found in S. griseus and S. avermitilis under the tested conditions.
Loss of SLP genes affects the microbial competition between Streptomyces lividans and fungi
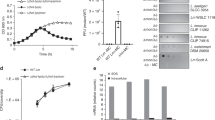
Next, we investigated the biological functions of SLPs using S. lividans as a model. It has long been known that bldA, a key regulator of SLP expression (Supplementary Fig. S1)17, is an essential factor in the life cycle of Streptomyces species, which encompasses the following developmental stages: spore, vegetative mycelia, and aerial mycelia formation. Many genes essential for these morphological differentiations are regulated by bldA and, therefore, the deletion of bldA leads to a “naked” phenotype, wherein aerial mycelia and spores are lacking17. In addition, bldA also acts as a trigger for secondary metabolite production; many gene clusters responsible for secondary metabolite production contain UUA codons, suggesting a pleiotropic role of bldA in the Streptomyces life cycle17,23,24. Given these observations, we first examined the effects of the deletion of the SLP gene on morphological differentiation and secondary metabolite production in S. lividans. However, in our experiments, the spore formation rates and the liquid chromatography/mass spectrometry profiles of the extractable metabolites of ΔslpS mutant were comparable with those of the TK23 strain. These results support those of the previous study, wherein the absence of any detectable difference in morphological differentiation and secondary metabolite production between M600 strain and SLP-deficient mutant of S. coelicolor was reported18,19. To further assess the phenotypic consequences of the deletion of SLP genes, we measured the growth of the TK23 S. lividans strain and two SLP-deficient strains (ΔslpS and ΔslpR). This revealed that slpR deletion leads to the fast-growing phenotype, whereas slpS deletion causes a slight increase in cell weight (Supplementary Fig. 2). Considering that the expression levels of SLP proteins were comparable to those of ribosomal proteins in S. coelicolor21, we believe that abolishing SLP expression allows the S. lividans strain to use more energy and resources for mycelial growth than the SLP-producing strain. In addition, to detect SLP expression and observe its distribution, we constructed an S. lividans strain expressing msfGFP-fused SlpS (SlpS-msfGFP). Microscopic analysis revealed that SlpS-msfGFP fluorescence is uniformly distributed in S. lividans colonies grown under standard conditions (Supplementary Fig. 3). These results are consistent with those of the previous study showing that SCO4253, a SlpS homolog in S. coelicolor, is expressed constantly during growth18,19. The abundant and constant expression of structural proteins during the growth phase has also been reported in the T6SSs of Gram-negative bacteria25, which may relate to their ecological importance. In addition, it has recently been proposed that some genes, including bldA-dependent ones, that are associated with the Streptomyces life cycle could mediate interactions between Streptomyces species and other organisms26,27. These observations led us to hypothesize that producing SLPs may have a significance in the biological interactions that Streptomyces species would constantly face in their natural settings. Given that Streptomyces species are often found in complex microbial communities in natural environments28, we speculated that the deletion of SLP genes lead to phenotypes that are disadvantageous to Streptomyces species in microbial competitions that could occur in such environments. For this reason, we co-cultured S. lividans and SLP-deficient mutants with various microorganisms and observed the colony morphologies. Among the tested microorganisms, S. cerevisiae and S. pombe were found to more severely invade the ΔslpS mutant colony than the TK23 strain of S. lividans (Fig. 3A,B). The slpS-complemented strain and ΔslpR mutant showed similar phenotypes to the TK23 strain and ΔslpS mutant, respectively, in the above competition assay, confirming the involvement of SLPs in the competitive interactions (Fig. 3A,B).
Deletion of SLP genes affects the microbial competition between S. lividans and fungi. Microscopic analyses of the microbial competition between S. lividans and fungi. (A) S. cerevisiae and (B) S. pombe were co-cultured with S. lividans on solid YES medium for 7 days. S. lividans and fungi were inoculated on the left and the right sides, respectively. White arrows indicate fungal invasion. The slpS-complemented strain was constructed by replacing the deleted slpS gene with intact slpS gene in its native chromosomal locus. In case of co-culture with S. cerevisiae, S. lividans and the fungus were cultured on a cellophane placed on the solid medium. Scale bars, 1 mm.
To gain more insights into the competition between S. lividans and fungi, we performed confocal laser scanning microscopy (CLSM) to visualize the colony boundaries. S. lividans and the fungi were visualized using Syto59 dye and GFP, respectively. The CLSM revealed that the colony boundary between the TK23 strain of S. lividans and S. pombe tends to be distorted, whereas distinct borders are formed between the ΔslpS mutant and the fungi (Fig. 4). In addition, the slpS-complemented strain showed similar phenotypes to the parental strain (Fig. 4). These results show that the outcomes of the microbial competitions between S. lividans and the fungi can be altered by the presence or absence of SLP genes.
Confocal laser scanning microscopy (CLSM) of the colony boundaries between S. lividans and fungi. Colony boundaries between S. lividans and fungi were observed by CLSM. (A) S. lividans strains were co-cultured with S. pombe (top) and S. cerevisiae (bottom) on a cellophane, and the bottom of the colony boundary was then observed by CLSM. A dotted arrow in the bottom panel indicates the observed region between the colonies. S. lividans and fungi were visualized using Syto59 dye staining (magenta) and GFP expression (green), respectively. Scale bars, 50 μm.
Differential expression patterns of SLP genes and antibiotic biosynthesis genes under co-culture conditions
To further confirm the involvement of SLPs in the interkingdom competition, we analyzed SLP expression in S. lividans under co-culture conditions with fungal competitors. In the co-culture condition with S. pombe, SlpS-msfGFP fluorescence was clearly observed at the colony boundary between S. lividans and the fungi (Fig. 5A).
Patterns of SLP expression and antibiotic production by S. lividans in response to microbial competitions. Microscopic and transcriptional analysis revealed the differential response of S. lividans to the competitive interactions. S. lividans expressing SlpS-msfGFP was co-cultured with (A) S. pombe and (B) B. subtilis 168. Scale bars, 2 mm. (C) Colony boundaries between B. subtilis and S. lividans (TK23 strain and ΔslpS mutant) were observed. Scale bars, 1 mm. Relative transcription levels of (D) slpS, (E) redD, and (F) cdaR were compared among the different culture conditions. The amounts of the transcripts were normalized to the transcription level of hrdB encoding RNA polymerase principal sigma factor B. All values and the bars represent the mean value ± S.D. for three independent cultures. P values were calculated by t test with Welch's correction.
Furthermore, for simultaneous observation of the CLSM findings of the colony boundary and SlpS expression, we constructed an S. lividans strain expressing SlpS-mScarletI, the fluorescence of which can be detected separately from that of the Syto59 dye and GFP. When this S. lividans strain was cultured with fungal competitors, the fluorescence of SlpS-mScarletI was detected at the colony boundary (Supplementary Fig. 4), clearly indicating that S. lividans expresses SLPs in the contact region between these microorganisms.
We were also interested in whether SLPs are expressed at the colony boundary between S. lividans and the bacterial competitor such as B. subtilis the competition with which were not altered by the presence or absence of SLPs in our experiment. The microscopic observation of the expression of the fluorescent protein-labelled SlpS under the co-culture condition with B. subtilis 168 revealed that SlpS-msfGFP fluorescence was absent at the colony boundary, whereas S. lividans accumulated red pigments at the colony boundary, which strongly indicates the co-culture-dependent induction of secondary metabolite production, including that of the red antibiotic prodiginines (RED)29,30,31 (Fig. 5B). In the competition assay with B. subtilis, the ΔslpS mutant of S. lividans showed similar RED production to that of the TK23 strain, suggesting that the absence of SLPs at the colony boundary does not affect RED production induced by the interbacterial competition (Fig. 5C).
Given these results, we speculated that S. lividans increases antibiotic production in response to the interbacterial competition while decreasing SLP expression. We thus compared the transcription levels of the structural gene of SLP and antibiotic biosynthetic genes under co-culture conditions with bacterial or fungal competitors. We co-cultured S. lividans with either S. pombe or B. subtilis and extracted total RNA to quantify the amount of the transcripts of slpS and redD, a gene encoding a pathway-specific activator protein for RED production in S. lividans32. Subsequent quantitative reverse transcription polymerase reaction (RT-qPCR) analysis revealed that the transcription level of slpS was approximately two times higher in the co-culture with S. pombe than in the single culture (Fig. 5D), indicating that SLP expression was induced in response to contact with the fungus, whereas the redD transcription levels were comparable between these culture conditions (Fig. 5E). In contrast to the co-culture conditions with the fungal competitor, the transcription level of slpS was rapidly decreased during co-culture with B. subtilis, which is consistent with the absence of SlpS-msfGFP fluorescence at the colony boundary between S. lividans and B. subtilis (Fig. 5B). In addition, significantly higher levels of redD transcription in the co-culture with B. subtilis (Fig. 5E) support the previous observation that the induction of RED production in S. lividans can be triggered by interbacterial competition (Fig. 5B)29,30,31.
Besides RED, S. lividans produces several antibiotics such as calcium-dependent antibiotic (CDA), a channel-forming antibiotic that is effective against various Gram-positive bacteria, including B. subtilis33,34. CDA production is regulated by cdaR encoding a pathway-specific activator protein CdaR35. The transcription levels of cdaR under the co-culture condition with B. subtilis showed similar tendency to that of redD (Fig. 5F), indicating that the co-culture with B. subtilis also triggered the upregulation of the biosynthetic genes responsible for CDA production. Moreover, although co-culture with B. subtilis showed these positive effects on the antibiotic production in S. lividans, co-culture with S. pombe did not alter the transcription level of cdaR (Fig. 5F). In addition, this tendency of the transcriptional responses of S. lividans to S. pombe was also observed in the co-culture with S. cerevisiae (Supplementary Fig. 5). Together with the results of the microscopic analysis (Fig. 5A,B), these transcriptional responses demonstrate that the patterns of SLP expression and antibiotic production in S. lividans can differ depending on its competitor species.
Discussion
In recent years, bioinformatic analyses have revealed that eCIS gene clusters are highly conserved among diverse bacterial classes, including Gram-negative and Gram-positive bacteria and archaea8,9. To date, the biological significance of eCIS has been established in only a few Gram-negative bacterial species. Anti-feeding prophage, an intensively studied model of eCIS, was discovered as a plasmid-encoded phage tail-like contractile injection system from Serratia entomophila, a causal agent of amber disease of the New Zealand grass grub Costelytra zealandica13. Further studies have revealed that anti-feeding prophage encoded by a gene cluster comprising 18 open reading frames is a key factor for eliciting amber disease in grass grub13. Photorhabdus virulence cassettes, produced by the symbiotic Photorhabdus species, are another model of eCIS with insecticidal activity14 and are chromosomally encoded in a pathogenicity island in which a diverse repertoire of virulence genes combating the hosts is encoded14. Conversely, metamorphosis-associated contractile structures from Pseudoalteromonas luteoviolacea represent eCISs with distinct structural properties and biological functions15. Metamorphosis-associated contractile structures were isolated from P. luteoviolacea, which exists in a symbiotic partnership with a marine tubeworm, and shown to be essential in the invertebrate’s metamorphosis15. Although these previous observations have demonstrated the versatile functions of eCISs in the interkingdom interactions between prokaryotes and eukaryotes, to the best of our knowledge, there has been no report elucidating the biological function of eCISs conserved in Gram-positive bacteria. One of the largest groups in which eCIS-related genes are widely conserved is Gram-positive actinomycetes, especially the Streptomyces species8,9. Since the first description by Kim et al. in 200519, it has been strongly suggested that eCIS-related genes confer selective advantages to Streptomyces species9,18,19. The importance of these eCIS-related genes of Streptomyces is also suggested by the fact that they are often encoded in the central region of their linear genomes where many of the essential genes are encoded26. However, the biological significance of these genes has so far remained unclear because of the absence of apparent phenotypic changes in the deletion mutants for these genes under standard laboratory conditions18,19. In the present study, we have shown that SLPs, which are eCIS-like nanostructures produced by Streptomyces, could confer ecological benefits to S. lividans in microbial competitions. Our results indicate that SLP production protects S. lividans colonies from fungal invasion.
Streptomyces species and fungi are ubiquitously found in polymicrobial communities such as soil, occupying a substantial niche within the densely populated environments28. In such environments wherein nutrient availability is often limited1,2, SLP-mediated defense against fungal invasion could be beneficial to Streptomyces species because the defensive role of producing SLPs may increase the possibility of Streptomyces species acquiring nutrients, forming spores, and ultimately occupying the ecological niches. Another example of recently reported competitive interactions between Streptomyces species and fungal competitors is an indirect interkingdom interaction that triggered the morphological transition of S. venezuelae36. In this interaction, glucose depletion by neighboring fungi triggered the exploratory growth of S. venezuelae; the S. venezuelae colony rapidly traversed both the biotic and abiotic surfaces36. Although this exploratory phenotype was observed in some Streptomyces isolates, it is notable that most of the model Streptomyces species, including S. lividans and S. coelicolor, do not exhibit the exploratory phenotype36. In addition, the transcription levels of the SLP-related genes conserved in S. venezuelae remain unchanged in the phenotypic transition induced by glucose limitation, indicating that SLP-related genes are not involved in the exploratory growth of S. venezuelae36. Therefore, this distinct mode of growth would be irrelevant to the SLP-mediated competition between the Streptomyces species and fungi that we found in this study. Although an underlying mechanism of SLP-mediated microbial interactions is under investigation, our findings would provide new insights into interkingdom competitions that occur between Streptomyces species and its eukaryotic competitors. Further, the constitutive SLP expression in the absence of the fungal competitors suggests that the expression of SLPs can be triggered in response to, in addition to the interaction with the fungal competitors, general environmental stimuli such as nutrient levels which could provoke bldA-dependent gene expressions17. In this view, producing SLP may be connected to adaptation to various environmental conditions and may thus have biological significance in a broad range of growth conditions besides the microbial competition. Applying growth conditions that closely mimic the natural settings of Streptomyces would facilitate further clarification of the biological roles of SLPs, and this will be our next challenge.
It has been established that competitive interactions between Streptomyces species and other bacteria could be mediated by antibiotics4,5,29,37. This has been exemplified by a co-culture model system of S. lividans and B. subtilis, in which S. lividans accumulates antibiotic RED upon contact with B. subtilis colonies, whereas B. subtilis produces bacillaene, which exhibits growth inhibitory activity against Streptomyces species4,29. In the present study, we found that SLP expression in S. lividans was decreased during co-culture with B. subtilis, whereas antibiotic production was significantly increased as previously reported4,29 (Fig. 5). Similar transcriptional responses in interbacterial competitions have been reported in the previous study; the study demonstrated that SLP gene expression is decreased in S. coelicolor under co-culture conditions with the bacterial competitor Myxococcus xanthus, whereas the expression of genes involved in antibiotic production was elevated5. These observations and our results suggest that the upregulation of the genes responsible for antibiotic production and eventual decrease of SLP expression may be common responses between these Streptomyces species and the prokaryotic competitors. However, it should be noted that the transcription level of slpS under co-culture conditions with B. subtilis was initially increased and then dramatically decreased within 3 h (Fig. 5D). This suggests that the initial expression of SLPs might be triggered by a mechanism involving a general response to competitor microorganisms, including both bacteria and fungi, although knowledge on such mechanisms is currently limited in Streptomyces species30,31. In addition, metabolic switching from SLP expression, which is likely to burden S. lividans by consuming considerable resources and energy (Supplementary Figs. 2 and 3)21, to antibiotic production might have occurred in the case of the competition with B. subtilis and possibly other bacterial competitors4,5,29. These implications of the recognition of other microorganisms by Streptomyces species and the differential responses to the microbial competitions would raise interesting questions regarding the behavior of Streptomyces species in nature.
In the present study, we demonstrate that Streptomyces species produce SLPs, that can potentially protect Streptomyces colonies from fungal invasion. Differential patterns of SLP expression and antibiotic production by Streptomyces species in response to microbial competition are suggestive of as-yet unidentified strategies of Streptomyces species to survive and thrive in diverse environments densely populated with various microorganisms. Further, our findings also provide insights into co-culture-based approaches aiming at the discovery of the novel traits of Streptomyces species, including the production of new antibiotics.
Methods
Strains and culture conditions
The strains used in this study are listed in Supplementary Table 1. Streptomyces species were cultured in Bennett's glucose (BeG) medium comprising 0.1% (w/v) yeast extract, 0.1% (w/v) meat extract, 0.2% (w/v) NZ amine, and 1% (w/v) glucose. The pH was adjusted to 7.2. For measurement of the growth curve, 105 viable spores were inoculated in 100 mL of liquid BeG medium and incubated at 30 °C with shaking at 180 rpm. The Streptomyces species and fungi were co-cultured in yeast extract with supplements (YES) medium comprising 0.5% (w/v) and 1% (w/v) glucose. The co-culture experiment was performed as follows: Streptomyces (104 viable spores) were inoculated onto solid YES medium and cultured at 30 °C for 20 h. The fungi were precultured in 10 mL liquid YES medium for 1 day and then inoculated (10 μL) onto a site adjacent to the precultured S. lividans colony. For co-culture with B. subtilis, solid BeG medium was used. The initial distance between the colonies was approximately 1–2 mm. The cultivation periods varied from 4 to 7 days depending on the co-culture conditions. If required, S. lividans and fungi were co-cultured on a cellophane placed onto a solid medium.
Genetic manipulations
Gene disruption of Streptomyces species was performed as follows. For S. lividans, the pK18mob plasmid was used as a conjugation vector; it was digested with EcoRI and HindIII and then fused with the amplified flanking regions (approximately 2 kbp each) of a gene of interest using In-Fusion (Clontech Laboratories Inc., CA, USA). The constructed plasmid was transformed into E. coli S17-1. The plasmid was then transferred from the transformed E. coli S17-1 to S. lividans by conjugation. Gene-disrupted mutants were obtained through homologous recombination events and consequent in-frame deletion of the targeted gene. Kanamycin (20 μg/mL) was used as a selection marker. For fusion of slpS with fluorescent proteins, the C-terminal region of slpS was amplified and fused with a flexible linker and codon-optimized msfgfp or mscarletI. The fused sequence was then introduced into the pK18mob plasmid, and the resultant plasmid was transferred to S. lividans as described above. Replacement of native slpS with slpS-msfgfp or slpS-mscarletI was completed through homologous recombination events. For gene disruption in S. albus, a pAT19 plasmid carrying an apramycin resistance cassette was used instead of pK18mob. E. coli ET12567 (pUB307) was used as a donor strain for conjugation. Each of the internal sequences of XNR_0535 and XNR_0530 was cloned into the pAT19 plasmid and then transferred to S. albus. Each gene was disrupted by a single homologous recombination event. The resultant mutants were obtained using apramycin (20 μg/mL) as a selection marker. Primers and plasmids used in this study are listed in Supplementary Table 2.
Extraction of SLPs
SLPs were extracted from Streptomyces mycelia as follows. Viable spores (105 per plate) were spread onto a cellophane placed on a BeG agar plate. After incubation at 30 °C for 3–5 days, the mycelia were scraped and resuspended in lysis buffer containing 50 mM Tris–HCl buffer (pH 7.5), 150 mM NaCl, 1 mg/mL lysozyme, 100 μg/mL DNase, 1% (v/v) Triton X-100, and a protease inhibitor cocktail. The resuspended solution was incubated at 37 °C for 2 h. After centrifugation at 15,000 × g for 10 min, the supernatant was ultracentrifuged at 200,000 × g for 60 min. The pellets were resuspended in resuspension buffer containing 50 mM Tris–HCl buffer (pH 7.5), 150 mM NaCl, and a protease inhibitor cocktail. The resultant solutions were subjected to further analysis. For TEM, the extracted SLPs were attached to thin carbon film-coated TEM grids (ALLIANCE Biosystems, Osaka, Japan) and washed with H2O. SLPs were then visualized via negative staining.
Microscopic analysis
CLSM was performed using LSM880 equipped with objective LD LCI Plan-Apochromat 25 × /0.8 Imm Corr DIC M27 (Carl Zeiss, Oberkochen, Germany). The mycelia of S. lividans were visualized as follows. S. lividans and fungi were co-cultured on a cellophane placed on solid YES medium. After cultivation, the cellophane was transferred to fresh YES medium containing 5 μg/mL Syto59 and subjected to CLSM. Note that fungal cells appear not to be stained by Syto59 dye, probably due to robust transporters pumping out the dye. The colony was observed using Axio Zoom (Carl Zeiss).
Bioinformatic analyses
The amino acid sequences of the eCIS tube proteins were obtained from database for eCIS9. Evolutionary analyses were conducted in MEGA X38. The tree was drawn to scale, with branch lengths in the same units of the evolutionary distances used to infer the phylogenetic tree. All ambiguous positions were removed for each sequence pair (pairwise deletion option). Bootstrap values were calculated from 500 replicates. Comparison and visualization of gene cluster similarity were performed using clinker, a Python-based tool, and clustermap.js, a companion JavaScript visualization library (https://github.com/gamcil/clinker)39.
RT-qPCR analysis
Transcriptional analysis of S. lividans was performed as follows. S. lividans, B. subtilis, and S. pombe were precultured in 10 mL liquid YES medium for 20 (B. subtilis) or 42 (S. lividans, S. pombe and S. cerevisiae) h. The precultured cells were then washed and resuspended in fresh YES medium. An equivalent volume (500 μL) of the resuspended solutions were mixed in sterilized 1.5-mL tubes and incubated at 30 °C. To note, wet weights of the inoculated cells were adjusted to 30–50 mg/tube for all culture conditions. After incubation, the tubes were centrifuged at 7000 × g for 10 min. Total RNA was extracted from the harvested cells using the RNeasy Mini kit (QIAGEN, Venlo, Netherlands).
RT-qPCR was performed as follows. The extracted RNA was treated with DNase and then purified via ethanol precipitation. The purified RNA was subjected to cDNA synthesis. The synthesized cDNA was then mixed with TB Green master mix (Takara Bio Inc., Shiga, Japan) and primers (0.2 μM each) just before the subsequent reaction. Reaction specificities were confirmed using melting curve analysis. The thermal profile of the qPCR was as follows (2-step PCR protocol): 95 °C for 3 min, 95 °C for 5 s, and 60 °C for 30 s (30 cycles). The standard curve for the calculation of relative abundance of each transcript was obtained using sequentially diluted genomic DNA (0.05–50 ng) as templates. Primers used are listed in Supplementary Table 2.
Data availability
The datasets supporting the current study are available from the corresponding author on request.
References
Hibbing, M. E., Fuqua, C., Parsek, M. R. & Peterson, S. B. Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15–25 (2010).
Stubbendieck, R. M. & Straight, P. D. Multifaceted interfaces of bacterial competition. J. Bacteriol. 198, 2145–2155 (2016).
Ghoul, M. & Mitri, S. The ecology and evolution of microbial competition. Trends Microbiol. 24, 833–845 (2016).
Straight, P. D., Fischbach, M. A., Walsh, C. T., Rudner, D. Z. & Kolter, R. A singular enzymatic megacomplex from Bacillus subtilis. Proc. Natl. Acad. Sci. USA 104, 305–310 (2007).
Lee, N. et al. Iron competition triggers antibiotic biosynthesis in Streptomyces coelicolor during coculture with Myxococcus xanthus. ISME J. 14, 1111–1124 (2020).
Russell, A. B. et al. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347 (2011).
Leiman, P. G. et al. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. USA 106, 4154–4159 (2009).
Sarris, P. F., Ladoukakis, E. D., Panopoulos, N. J. & Scoulica, E. V. A phage tail-delivered element with wide distribution among both prokaryotic domains: a comparative genomic and phylogenetic study. Genome Biol. Evol. 6, 1739–1747 (2014).
Chen, L. et al. Genome-wide identification and characterization of a superfamily of bacterial extracellular contractile injection systems. Cell Rep. 29, 511–521 (2019).
Desfosses, A. et al. Atomic structures of an entire contractile injection system in both the extended and contracted states. Nat. Microbiol. 4, 1885–1894 (2019).
Jiang, F. et al. Cryo-EM structure and assembly of an extracellular contractile injection system. Cell 177, 370–383 (2019).
Shikuma, N. J., Antoshechkin, I., Medeiros, J. M., Pilhofer, M. & Newman, D. K. Stepwise metamorphosis of the tubeworm Hydroides elegans is mediated by a bacterial inducer and MAPK signaling. Proc. Natl. Acad. Sci. USA 113, 10097–10102 (2016).
Hurst, M. R. H., Glare, T. R. & Jackson, T. A. Cloning Serratia entomophila antifeeding genes—a putative defective prophage active against the grass grub Costelytra zealandica. J. Bacteriol. 186, 5116–5128 (2004).
Yang, G., Dowling, A. J., Gerike, U., Ffrench-Constant, R. H. & Waterfield, N. R. Photorhabdus virulence cassettes confer injectable insecticidal activity against the wax moth. J. Bacteriol. 188, 2254–2261 (2006).
Shikuma, N. J. et al. Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like structures. Science 343, 529–533 (2014).
Merrick, M. J. A morphological and genetic mapping study of bald colony mutants of Streptomyces coelicolor. J. Gen. Microbiol. 96, 299–315 (1976).
Hackl, S. & Bechthold, A. The gene bldA, a regulator of morphological differentiation and antibiotic production in Streptomyces. Arch. Pharm. Chem. Life Sci. 348, 455–462 (2015).
Hesketh, A. et al. New pleiotropic effects of eliminating a rare tRNA from Streptomyces coelicolor, revealed by combined proteomic and transcriptomic analysis of liquid cultures. BMC Genomics 8, 261 (2007).
Kim, D. W., Chater, K. F., Lee, K. J. & Hesketh, A. Effects of growth and the developmentally significant bldA-specified tRNA on the membrane-associated proteome of Streptomyces coelicolor. Microbiology 151, 2707–2720 (2005).
Jayapal, K. P. et al. Multitagging proteomic strategy to estimate protein turnover rates in dynamic systems. J. Proteome Res. 9, 2087–2097 (2010).
Thomas, L. et al. Metabolic switches and adaptation deduced from the proteomes of Streptomyces coelicolor wild type and phoP mutant grown in batch culture. Mol. Cell. Proteomics 11(2) (2012).
Arndt, D. et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21 (2016).
Higo, A., Horinouchi, S. & Ohnishi, Y. Strict regulation of morphological differentiation and secondary metabolism by a positive feedback loop between two global regulators AdpA and BldA in Streptomyces griseus. Mol. Microbiol. 81, 1607–1622 (2011).
Li, W. et al. A genetic and bioinformatic analysis of Streptomyces coelicolor genes containing TTA codons, possible targets for regulation by a developmentally significant tRNA. FEMS Microbiol. Lett. 266, 20–28 (2007).
Lin, L., Lezan, E., Schmidt, A. & Basler, M. Abundance of bacterial type VI secretion system components measured by targeted proteomics. Nat. Commun. 10, 2584 (2019).
Zhang, Z. et al. Antibiotic production in Streptomyces is organized by a division of labor through terminal genomic differentiation. Sci. Adv. 6, eaay5781 (2020).
Becher, P. G. et al. Developmentally regulated volatiles geosmin and 2-methylisoborneol attract at soil arthropod to Streptomyces bacteria promoting spore dispersal. Nat. Microbiol. 5, 821–829 (2020).
Seipke, R. F., Kaltenpoth, M. & Hutchings, M. I. Streptomyces as symbionts: an emerging and widespread theme?. FEMS Microbiol. Rev. 36, 862–876 (2012).
Vargas-Bautista, C., Rahlwes, K. & Straight, P. Bacterial competition reveals differential regulation of the pks genes by Bacillus subtilis. J. Bacteriol. 196, 717–728 (2014).
Asamizu, S., Ozaki, T., Teramoto, K., Satoh, K. & Onaka, H. Killing of mycolic acid-containing bacteria aborted induction of antibiotic production by Streptomyces in combined-culture. PLoS ONE 10, e0142372 (2015).
Onaka, H., Mori, Y., Igarashi, Y. & Furumai, T. Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl. Environ. Microbiol. 77, 400–406 (2011).
White, J. & Bibb, M. bldA dependence of undecylprodigiosin production in Streptomyces coelicolor A3(2) involves a pathway-specific regulatory cascade. J. Bacteriol. 179, 627–633 (1997).
Hopwood, D. A. & Wright, H. M. CDA is a new chromosomally-determined antibiotic from Streptomyces coelicolor A3(2). J. Gen. Microbiol. 129, 3575–3579 (1983).
Lakey, J. H., Lea, E. J. A., Rudd, B. A. M., Wright, H. M. & Hopwood, D. A. A new channel-forming antibiotic from Streptomyces coelicolor A3(2) which requires calcium for its activity. J. Gen. Microbiol. 129, 3565–3573 (1983).
Ryding, N. J., Anderson, T. B. & Champness, W. C. Regulation of the Streptomyces coelicolor calcium-dependent antibiotic by absA, encoding a cluster-linked two-component system. J. Bacteriol. 184, 794–805 (2002).
Jones, S. E. et al. Streptomyces exploration is triggered by fungal interactions and volatile signals. Elife 6, e21738 (2017).
Nishiyama, T., Hashimoto, Y., Kusakabe, H., Kumano, T. & Kobayashi, M. Natural low-molecular mass organic compounds with oxidase activity as organocatalysts. Proc. Natl. Acad. Sci. USA 111, 17152–17157 (2014).
Stecher, G., Tamura, K. & Kumar, S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 37, 1237–1239 (2020).
Gilchrist, C. L. M. and Chooi, Y. H. clinker & clustermap.js: automatic generation of gene cluster comparison figures. Bioinformatics btab007 (2020).
Acknowledgements
T. N. was supported by a Grant-in-aid for Scientific Research from the Japanese Society for the Promotion of Science (19K15726 and 20J01208). This research was supported in part by a grant-in-aid from Amano Enzyme Inc. (to H. O. and S. A.) and the Japan Science and Technology Agency (to N. N.; ERATO project JPMJER1502). M. T. was supported by Suntory Rising Stars Encouragement Program in Life Sciences (SunRiSE). We thank Dr. Shinichi Nishimura (The University of Tokyo, Japan) and Dr. Akihisa Matsuyama (RIKEN, Japan; The University of Tokyo) for providing the S. pombe strain. We thank Prof. Yasuo Ohnishi, Prof. Yohei Katsuyama, and Dr. Takeaki Tezuka (The University of Tokyo) for their valuable comments on our results.
Author information
Authors and Affiliations
Contributions
T. N. and T. Y. performed the microscopic analyses. T. N. performed all other experiments. T. N. designed the study. T. N. and T. Y. analyzed the data. All authors discussed the results. T. N., T. Y., M. T., and H. O. drafted the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nagakubo, T., Yamamoto, T., Asamizu, S. et al. Phage tail-like nanostructures affect microbial interactions between Streptomyces and fungi. Sci Rep 11, 20116 (2021). https://doi.org/10.1038/s41598-021-99490-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-99490-8
This article is cited by
-
Cytoplasmic contractile injection systems mediate cell death in Streptomyces
Nature Microbiology (2023)
-
A contractile injection system is required for developmentally regulated cell death in Streptomyces coelicolor
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.