Abstract
Endemic Burkitt lymphoma (eBL) is an aggressive pediatric B cell lymphoma, common in Equatorial Africa. Co-infections with Epstein-Barr virus (EBV) and Plasmodium falciparum, coupled with c-myc translocation are involved in eBL etiology. Infection-induced immune evasion mechanisms to avoid T cell cytotoxicity may increase the role of Natural killer (NK) cells in anti-tumor immunosurveillance. Killer immunoglobulin-like receptor (KIR) genes on NK cells exhibit genotypic and allelic variations and are associated with susceptibility to diseases and malignancies. However, their role in eBL pathogenesis remains undefined. This retrospective study genotyped sixteen KIR genes and compared their frequencies in eBL patients (n = 104) and healthy geographically-matched children (n = 104) using sequence-specific primers polymerase chain reaction (SSP-PCR) technique. The relationship between KIR polymorphisms with EBV loads and eBL pathogenesis was investigated. Possession of ≥ 4 activating KIRs predisposed individuals to eBL (OR = 3.340; 95% CI 1.530–7.825; p = 0.004). High EBV levels were observed in Bx haplogroup (p = 0.016) and AB genotypes (p = 0.042) relative to AA haplogroup and AA genotype respectively, in eBL patients but not in healthy controls. Our results suggest that KIR-mediated NK cell stimulation could mute EBV control, contributing to eBL pathogenesis.
Similar content being viewed by others
Introduction
Endemic Burkitt lymphoma (eBL) is the quintessential Epstein-Barr Virus (EBV)-associated B cell malignancy in pediatric patients within Africa and Papua New Guinea1. In Africa, eBL has the highest incidence in areas where Plasmodium falciparum (Pf) malaria is common, hence repeated interaction of Pf-infected red blood cells with EBV-infected B cells is postulated to result in eBL oncogenesis1, 2. The proposed mechanism of eBL development involves a combination of activation-induced cytidine deaminase (AID)-associated c-myc chromosomal translocation, modulation of host T cell immunity to EBV antigens, monoclonal expansion of B cells infected with EBV, and reactivation of EBV, resulting in increased viremia3. Cells infected with EBV down-regulate the expression of human leukocyte antigen (HLA) to evade recognition by HLA-restricted cytotoxic CD8 + T cells4. However, this immune evasion mechanism, i.e. ‘missing-self’ should render them susceptible to killing by natural killer (NK) cells4. NK cells constitute the body's first line of defense against viral infections and tumor cells5. They are identified by the expression of CD56, a neural cell adhesion molecule 1 (NCAM-1) belonging to the immunoglobulin supergene family6. This molecule mediates cell to cell interactions and its surface expression levels vary with cell maturation6. Consequently, there are two major NK cell populations: CD56bright are mainly cytokine-producing7, while CD56dim acquire additional CD16 and killer immunoglobulin-like receptors (KIRs) receptors, which enhance their cytolytic activities7, 8. Other studies have reported accumulation of CD56 negative CD16 positive NK cell subsets in eBL patients9, and in HIV-infected individuals10. NK cell anti-viral and anti-tumor activities are partly regulated by inhibitory (iKIRs) and activating (aKIRs) KIRs, which are also expressed by some TCR-γδ, CD8+, and CD4+ T cells11,12,13. KIRs interact with various HLA class I ligands on target cells14. Tumor cells lacking ligands for inhibitory and activating KIRs do not stimulate NK cells response15. Additionally, NK cells do not kill healthy cells when only the inhibitory receptors are ligated to HLA-I ligands on the target cells, since there is no activating signal generated15. Down-regulation of HLA-I in target cells by viral infections or neoplastic transformation results in a lack of ligation of inhibitory NK cell receptors to their ligands, hence the absence of NK cell inhibition. Instead, only the activating NK cell receptors are ligated to activating ligands, resulting in NK cell stimulation to kill target cells16. The outcome of the interaction of NK cells with tumor cells containing ligands for both inhibitory and activating receptors depends on the balance of the strength of signals generated15.
The KIR gene family comprises rapidly evolving genes present in all primates17. The genes contain two (2D) or three (3D) domains in the extracellular region, with a short (S) or a long (L) cytoplasmic tail18. The KIR genes 2DL1, 2DL2/2DL3, 2DL5, 3DL1, 3DL2, and 3DL3 have a long tail with an inhibitory motif. Short cytoplasmic tailed KIRs have activating motifs and include 2DS1, 2DS2, 2DS4, 2DS3/2DS5, and 3DS118. KIR2DL4 is the only long-tailed receptor with both inhibitory and activating motifs19. There are two pseudogenes, 2DP1 and 3DP120 (Fig. 1). These genes are arranged in a head-to-tail order in the long arm of chromosome 19 (19q13.4), within the Leukocyte Receptor Complex (LRC)21. Each KIR gene is 10–16 kb in length, with a 2 kb sequence separating each gene pair, except a 14 kb sequence that occurs upstream of KIR2DL421. The expression of KIR genes varies between NK cell subsets and is controlled by four types of promoters22. CD56dim NK cells express all KIR genes except 3DL322. 2DL4 occurs on both CD56bright and -dim NK cells in a non-variegated manner22. Some KIR genes demonstrate variations in their sequences, for example, KIR2DS4 has a 22 base pair (bp) deletion in the second extracellular domain which results in a non-functional gene21. A deletion of one base-pair in exon 4 of KIR2DP1 introduces a stop codon, resulting in a pseudogene23. Another pseudogene, KIR3DP1 has a deletion of 1.5 kb that removes exon 2. There are no transcripts for the 2 pseudogenes23. KIR2DL5 has two variants A and B, encoded by different loci24. 2DL5B is in the centromeric region, while 2DL5A occurs in the telomeric region25.
Polymorphisms within the KIR locus result from gene content, allelic, and copy number variations26. Based on gene content and copy number, KIRs are grouped into inhibitory haplotype A and activating haplotype B27. The haplotypes are further subdivided into AA and Bx genotypes, where x can be either A or B28. There are more than 500 different Bx groups in the database (http://www.allelefrequencies.net)28. KIR genotype AA is homozygous for the haplotype A and contains 3DL3, 2DL3, 2DP1, 2DL1, 3DP1, 2DL4, 3DL1, 2DS4, and 3DL2 genes21. Activator haplotype B has a variable number of activating KIRs, and comprise of 2DS2, 2DL2, 2DL5B, 2DL1, 2DP1, 3DP1, 3DL3, 2DL4, 3DS1, 2DL5A, 2DS3/2DS5, 2DS1, 2DS4, 2DS3/2DS5 and 3DL2 genes21. This haplotype has a Bx group containing one (AB heterozygous) or two (BB homozygous) genotypes. Genotype BB does not have one or more of the group A KIR genes. All the remaining genotypes in haplotype B are defined as AB29. 3DL3, 3DL2, 2DL4, and 3DP1 are framework genes, hence they appear in all haplotypes23. KIR haplotypes are split into centromeric A or B (cA, cB) and telomeric A or B (tA, tB) halves30, 31. Both cA, cB and tA, tB regions exhibit an even balance in East Africa population32. Classification of KIR based on presence/absence of a gene generates eight telomeric regions (tA01, tB01, tB02, tB03, tB04, tB05, tB06 and tB07) and nine centromeric regions (cA01, cA02, cA03, cB01, cB02, cB03, cB04, cB05 and cB06)33. KIR2DL5, 2DS5, and 2DS3 are duplicated and can occur in centromeric and/or telomeric locations34. Genes occurring in different regions of the KIR complex may undergo homologous recombination, resulting in expanded and contracted haplotypes34, 35 The B content score is the sum of cenB and/or telB motifs in each genotype36. The Bx group can be classified further into four subsets, by considering two gene clusters; T4, containing KIR2DL5-3DS1-2DS1-2DS5 genes, and C4, which has KIR2DL2-2DS2-2DS3-2DL5 genes. The C4T4 contains both C4 and T4 genes, while the C4Tx subset has C4 but lacks T4 genes. CxT4 lacks C4 genes, thus it contains T4 genes. The absence of both C4 and T4 genes results in CxTx subset29, 37.
Studies have suggested that specific KIRs influence the generation of either inhibitory versus activating signals. A balance between these signals determines whether NK cells bypass or kill viral-infected or tumor cells38. Consequently, these signals can influence an individual's susceptibility to diseases and malignancies16, 30, 39, 40. The presence of certain KIRs has been associated with cancer pathogenesis. For instance, an increased number of activating KIRs predispose individuals to EBV-related nasopharyngeal carcinoma (NPC)16, whereas the presence of genotype B, which mainly contains activating KIRs is associated with gastric cancer lesions30. In contrast, the Bx haplogroup protects against colorectal adenocarcinoma41. However, there is little understanding of the impact of KIR polymorphisms on eBL pathogenesis. Therefore, to improve our understanding of how KIR genes may contribute to eBL pathology, we performed KIR genotyping using commercially available kits and analyzed the haplotype, genotypes, centromere-telomere regions, Bx subsets, and B score contents in eBL patients and healthy controls. Given the strong link between EBV and eBL42, we further evaluated the association of haplogroups AA/Bx and genotypes AA, AB, and BB with EBV loads, to determine viral control.
Results
KIR genes
To characterize the frequencies of KIR genes in the study population, we genotyped the genes responsible for inhibitory signals (2DL1, 2DL2/2DL3, 2DL5, 3DL1), activating signals (2DS1, 2DS2, 2DS4, 2DS3/2DS5, and 3DS1), the framework and pseudogenes (2DL4, 3DL2, 3DL3, 2DP1 and 3DP1) from genomic DNA using sequence-specific primers polymerase chain reaction (SSP-PCR) technique. KIR genotypes were classified based on the presence or lack of each gene locus and were analyzed to determine differences in their frequencies between eBL patients and healthy controls (HC). The genes KIR3DP1, KIR2DP1, KIR2DL1, KIR2DL4, KIR3DL2 and KIR3DL3 occurred at a frequency ≥ 99% and were excluded from the association analysis. The KIR genes were not statistically different between the study groups (Table 1).
KIR haplotypes and genotypes
In the studied population, the haplotypes A and B occurred at frequencies of 56.7% vs. 60.6% and 43.3% vs 39.4% in eBL patients and HC respectively. The haplotypes were grouped into haplogroup AA (27.9% vs. 34.6%) and Bx (72.1% vs. 65.4%) for eBL patients and HC respectively (Table 2). There were 35 different haplogroups in the study population, based on the allele frequencies database (http://www.allelefrequencies.net)28 (Fig. 2). Out of these, 15 were identified in both cases and controls, fourteen had frequencies > 1.0%; representing 88.5% of the population, while eighteen had frequencies > 1.0% representing 94.2% of the healthy controls and eBL patients respectively. The remaining haplogroups (17 in eBL and 21 in HC) were rare, with frequencies ≤ 1.0%. The haplogroups were subdivided further into genotypes AA, AB, or BB according to the gene content. Among 104 eBL patients, 29 were genotypes AA (27.9%), 60 were AB (57.7%) while 15 were BB (14.4%). All the AA genotypes had ID 1. Among 104 HC, 36 were genotypes AA (34.6%), 54 were AB (51.9%) while 14 were BB (13.5%). The distribution of the KIR genotypes among the study groups was not statistically significant (Table 2).
The occurrence of KIR genotypes in the study population. Thirty-five different KIR genotypes were observed in the 208 persons. The genotypes differed from each other by the presence of (black box) or absence (open box) of KIR genes. The variants KIR2DL5A and 2DL5B were considered as KIR2DL5 while KIR2DS4 mutant and 2DS4 full length were considered as KIR2DS4, we analyzed a total of 16 KIR genes. The data are expressed as percentage frequency, obtained by dividing the number of individuals having the genotype by the number of individuals in the studied group. Genotype ID reference numbers were acquired from the KIR genotype database (http://www.allelefrequencies.net).
KIR centromeric and telomeric distribution
The KIR gene contents vary in the centromeric and telomeric regions. To investigate these differences in our study population, genotypes AA and Bx were grouped into centromeric and telomeric contents31. A total of 6 centromeric and 2 telomeric genetic regions were reported. There were no significant differences in these regions when comparing eBL patients with the control group (Supplementary Table S1).
KIR B score
To evaluate the involvement of B motifs in eBL development, the AA and Bx genotypes were investigated according to the distribution of B content in the centromeric (cB) and telomeric (tB) regions. A score of zero was more common in healthy controls while a score of one was frequent in eBL patients (44.2% vs. 38.5 and 43.3 vs. 50.0% respectively) (Supplementary Table S2). However, the observed differences were not statistically significant.
Bx Subsets and the number of activating KIRs
We observed that all the four Bx subsets (C4T4, CxT4, C4Tx, CxTx) were present in the study population (Table 2). C4T4 had the least frequency, with no representation among the healthy individuals. CxTx had the highest frequency in both eBL patients and healthy controls. Further, in order to investigate possible differences in the number of KIRs, we compared the iKIRs and aKIRs in cases and controls. There were an increased proportion of eBL patients with ≥ 4 aKIRs relative to HC.
KIR haplotypes, genotypes, and EBV Viral loads
We compared EBV viral loads and observed significantly higher viremia in children with eBL (median 6496.571 EBV copies/μg of DNA) compared to healthy children from the malaria holoendemic region (median 202.697 EBV copies/μg of DNA) (p-value <0.0001). Next, we investigated whether the EBV loads differed between the haplotypes and genotypes. Considering the KIR haplotypes and genotypes, we observed significant differences in EBV load in eBL patients but not in healthy controls (Fig. 3).
EBV load stratified by AA/Bx haplogroups and AA, AB, and BB genotypes. The EBV levels were compared for eBL patients (n = 93), and healthy controls (n = 80) after stratification by AA/Bx haplogroups (A,B) and AA, AB, and BB genotypes (C,D). Significant differences in EBV viral loads were associated with AA/Bx haplogroups and AA/AB genotypes in eBL patients but not in healthy controls; based on Mann–Whitney test and one way Kruskal–Wallis statistic respectively. The p-value in C was statistically significant; hence pair-wise comparisons were assessed by Dunn's test. (*p < 0.05). ns = not significant.
Discussion
KIR gene polymorphisms predispose individuals to various malignancies associated with viruses16. However, few studies have evaluated the role of such polymorphisms in eBL etiology. To address this issue, we evaluated the association of KIR genes with eBL development. The most common KIR genotype in our study population was homozygous A, with the genotype id AA1. This genotype has previously been shown to be the most frequent in all worldwide populations, including Africans44,45,46. Interestingly, while its frequency in healthy controls was consistent with the expected frequency in African populations (35.6%)45, the representation was lower in eBL patients (27.9%). However, such variations have been reported in a few African populations, from 12.0% in the Xhosa population of South Africa, 28.1% in the Ugandan population, and 42.0% in Senegal44, 47, 48. In our study, the Bx genotype was highly variable, with a frequency range of 0.0–10.6%. This genotype consists of two haplotypes; AB and BB. Most of the study participants were heterozygous AB, and there was a very low frequency of homozygous BB. The considerable diversity for Bx but not AA genotypes may be a result of copy number variation33 due to selection pressure from environmental, climatic, chronic, and infectious diseases that have prevailed in our study population for many years40. Recently, malaria has been shown to drive selection for this haplotype in a Ugandan population49. In our study, there were more eBL patients carrying ≥ 4 aKIRs compared to healthy controls; suggesting that individuals with ≥ 4 aKIRs may have a high risk of developing eBL. Consistent with our findings, an increased number of aKIRs predispose individuals to colorectal adenocarcinoma, human papillomavirus-associated cervical cancer and EBV-associated nasopharyngeal carcinoma16, 39, 50. The role of the number of aKIRs in the etiology of cancers is explained by two hypotheses39. First, an increased number may protect individuals against cancers, due to enhanced cytolysis of tumor cells, resulting from increased NK cell activation30. In contrast, increased immune activation of NK cells by aKIRs may cause non-specific inflammatory responses, such as oxidative DNA damage16, 30. Such responses may increase the risk of cancer development30. Therefore, considering the second hypothesis, our findings raise a possibility that an increasing number of aKIRs coupled with repeated infections with Plasmodium falciparum in our study population51 could be associated with increased NK cell activation resulting in inflammation-associated oncogenesis.
The study participants had a higher frequency of centromeric B region and a lower frequency of telomeric B region. Similar observations were reported in a Ugandan population44. Generally, cenB region is common in the African population relative to telB region44. The number of B motifs in the centromere and telomere regions influences NK cell activation30. Subsequently, the B motif is associated with disease outcome30. In this study, we evaluated how the number of KIR B gene motifs of centromeric or telomeric origin influences eBL development. There were no significant differences in the B score when comparing eBL patients with the control group.
Previous studies have reported that children living in malaria holoendemic areas experience primary EBV infection at an early age compared to children residing in areas with lower incidences of malaria52. In addition, repeated exposure to malaria is associated with poor EBV control, hence higher viremia53. Consistent with these findings, we observed that eBL patients had higher median EBV loads relative to healthy controls (6496.571 versus 202.697 EBV copies/μg of DNA), respectively, p-value < 0.0001). The EBV levels were significantly different when considering the KIR haplogroup and genotypes in children with eBL but not in healthy controls, with higher EBV loads observed in Bx relative to AA haplogroup. Considering the genotypes, EBV load differed between AA and AB. NK cells are essential in the control of infections associated with viruses54. Their subsets expand upon infection with herpes viruses55, and the proliferation positively correlates with EBV viral loads55. Individuals deficient in NK cells are predisposed to herpes viruses-associated infections56. The persistence of viruses in an individual may cause chronic recruitment and activation of NK cells, up to when in some individuals; the NK cell activation is deregulated. Therefore, increased viral load in Bx relative to AA haplogroup in eBL could be related to continuous stimulation and subsequent loss of NK cell control of EBV. A previous study reported the accumulation of dysfunctional CD56 negative CD16 positive subset of NK cells in eBL patients9. Consistent with these findings, our results raise the possibility that activation of NK cells that are mediated by KIRs may impair NK cell functions in our study population9. Further studies are required to confirm this observation and the role of KIR-expressing T cells11,12,13.
A limitation of our study was the small sample size and convenience sampling of healthy controls which led to them being younger than eBL cases. However, since KIR genotypes do not change with age, we don’t believe this biased our findings. In addition, our conclusion is restricted by a lack of HLA ligands data, as KIR/HLA combinations influence NK cell activity. As KIRs can vary at the gene or allele level, we only investigated the presence or absence of each KIR gene, and we could not evaluate allelic and copy number variations that can impact NK cell functions. A previous study reported decreased expression of KIR2DL1/S1 inhibitory/activation marker and increased expression of KIR3DL1 in children exposed to malaria and eBL patients relative to healthy children9, hence future studies will need to assess the mechanistic implications of KIR proteins and gene expression profiles in eBL pathogenesis. We acknowledge that whereas our findings suggest a possible association of the Bx haplogroup and increased number of aKIRs with EBV load and eBL pathogenesis respectively, there is a need for a larger validation cohort and functional studies to confirm their biological relevance.
Materials and methods
Study site and subjects
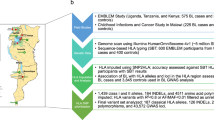
The study enrolled 208 children; 104 eBL patients, and 104 healthy individuals. Patients with eBL were children aged 0–14 years old, who were enrolled at Jaramogi Oginga Odinga Teaching and Referral Hospital (JOOTRH), located in Kisumu County, western Kenya. JOOTRH is one of the two regional referral centers for childhood cancer cases in western Kenya. Morphologic diagnosis of eBL was performed by staining fine-needle aspirates (FNA) with Giemsa/May-Grünwald and observed under a microscope57. The control group consisted of convenience samples selected from healthy non-eBL children, aged 0–12 years old, living in the same malaria-holoendemic regions of western Kenya as the eBL patients. Convenience sampling did not affect our results since KIR genotypes do not differ by age.
DNA extraction and KIR genotyping
Genomic DNA was isolated from 200 μl of blood using Qiagen QIAamp DNA Mini Kit (Valencia, CA, USA), following the manufacturer’s instructions, and frozen at − 20 °C until genotyping. The samples were analyzed for KIR gene content58. A commercially available KIR genotyping sequence-specific Primers (SSP) kit (Miltenyi, Biotec, Inc, Germany), was used to test for the presence or absence of KIRs generating inhibitory signals (KIR2DL1, 2DL2, 2DL3, 2DL5A, 2DL5B, 2DL5 (A and B) 3DL1), activating KIRs (2DS1, 2DS2, 2DS3, 2DS4del, 2DS4ins, 2DS5, and 3DS1) and the framework and pseudogenes (2DL4, 3DL2, 3DL3, 2DP1 and 3DP1) following the manufacturer’s recommendations. The amplified sequences were examined by electrophoresis in 2% agarose gel stained with SYBR Safe (Invitrogen, Burlington, ON, Canada) and visualized on a UV transilluminator using a gel documentation system (ChemiDoc, BioRad) for the presence or lack of amplicons specific to each gene, according to the manufacturer’s instructions (See full-length gel in Supplementary Fig. 1 online).
Definitions for KIR gene content polymorphisms
KIR polymorphisms were analyzed by determining the presence or absence of 16 KIRs genes; 2 pseudogenes, 8 inhibitory, and 6 activating KIR genes. The KIR haplotypes were classified into AA (inhibitory) and Bx (activator), where x can be A or B. The homozygote AA genotype was defined by the absence of KIR2DL2, 2DL5 (2DL5A and B, 2DL5A, 2DL5B) 2DS1, 2DS2, 2DS3, 2DS5, and 3DS1 genes. Individuals in the Bx genotype contained at least one of the genes above28. The genes 2DL4, 3DP1, 3DL2, and 3DL3 are framework genes23. The genotypes AA and Bx were evaluated according to the distribution of the centromeric and telomeric genes and the B content score as previously reported31.
EBV Load
Epstein-Barr virus load was determined by quantitative Polymerase Chain Reaction (qPCR)59. Briefly, amplification of genomic DNA was done in a Bio-Rad CFX96 Real-Time System with C1000 Thermo Cycler base for the primers and probes (BioRad Laboratories, Hercules, CA), following the manufacturer’s instructions. The PCR amplification conditions were: 180 s at 95 °C, 10 s at 95 °C, 30 s at 63.5 °C- plate read, 10 s at 95 °C (39 times).
Statistical analysis
The frequencies of KIR genes, haplotypes, genotypes, B score, and centromere-telomere gene content in eBL patients were compared with the healthy controls. Differences between KIR genes and EBV loads were assessed by Fisher’s exact test using Graphpad Prism version 8.0.2 software (GraphPad Software, La Jolla, CA). Comparisons of log-transformed EBV load between the genotypes were performed using the Mann–Whitney test and one way Kruskal–Wallis statistic. When the p-value in the Kruskal–Wallis test was statistically significant, pair-wise comparisons were assessed by Dunn's test. Multivariable logistic regression analyses were performed in R, controlling for age and sex as variables influencing the risk of eBL etiology. The statistical significance of associations was assessed using odds ratios (OR) with 95% confidence intervals (CI). A p ≤ 0.05 was considered significant.
Ethical approval
This research was approved by the Scientific and Ethical Review Unit (SERU) at the Kenya Medical Research Institute (KEMRI), and the Institutional Review Board at the University of Massachusetts Medical School (UMMS), Worcester, USA. All experiments were performed in accordance with relevant guidelines and regulations. Study participants were informed about the study and since they were all below 18 years, the parent and/or legal guardian provided written informed consent, before enrollment. In addition, children aged 13 years and above provided assent as per the requirements of the local IRB.
Data availability
The datasets evaluated in this study are available in supplementary data online.
References
Velavan, T. P. Epstein-Barr virus, malaria and endemic Burkitt lymphoma. EBioMedicine 39, 13–14 (2019).
Kafuko, G. & Burkitt, D. P. Burkitt’s lymphoma and malaria. Int. J. Cancer 6, 1–9 (1970).
Orem, J., Mbidde, E. K., Lambert, B., De Sanjose, S. & Weiderpass, E. Burkitt’s lymphoma in Africa, a review of the epidemiology and etiology. Afr. Health Sci. 7, 166–175 (2007).
Lodoen, M. B. & Lanier, L. L. Viral modulation of NK cell immunity. Nat. Rev. Microbiol. 3, 59 (2005).
Bhat, R. & Rommelaere, J. Emerging role of natural killer cells in oncolytic virotherapy. ImmunoTargets Therapy 4, 65 (2015).
Lanier, L. et al. Molecular and functional analysis of human natural killer cell-associated neural cell adhesion molecule (N-CAM/CD56). 146, 4421–4426 (1991).
Cooper, M. A., Fehniger, T. A. & Caligiuri, M. A. The biology of human natural killer-cell subsets. Trends Immunol. 22, 633–640 (2001).
Jacobs, R. et al. CD56bright cells differ in their KIR repertoire and cytotoxic features from CD56dim NK cells. Eur. J. Immunol. 31, 3121–3126 (2001).
Forconi, C. S. et al. Poorly cytotoxic terminally differentiated CD56negCD16pos NK cells accumulate in Kenyan children with Burkitt lymphomas. Blood Adv. 2, 1101–1114 (2018).
Hu, P.-F. et al. Natural killer cell immunodeficiency in HIV disease is manifest by profoundly decreased numbers of CD16+ CD56+ cells and expansion of a population of CD16dimCD56-cells with low lytic activity. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 10, 331–340 (1995).
Nakajima, T. et al. De novo expression of killer immunoglobulin-like receptors and signaling proteins regulates the cytotoxic function of CD4 T cells in acute coronary syndromes. Circ. Res. 93, 106–113 (2003).
Battistini, L. et al. Phenotypic and cytokine analysis of human peripheral blood gamma delta T cells expressing NK cell receptors. J. Immunol. 159, 3723–3730 (1997).
Mingari, M. C. et al. Human CD8+ T lymphocyte subsets that express HLA class I-specific inhibitory receptors represent oligoclonally or monoclonally expanded cell populations. Proc. Natl. Acad. Sci. 93, 12433–12438 (1996).
Wagtmann, N., Rajagopalan, S., Winter, C. C., Peruui, M. & Long, E. O. Killer cell inhibitory receptors specific for HLA-C and HLA-B identified by direct binding and by functional transfer. Immunity 3, 801–809 (1995).
Lanier, L. L. NK cell recognition. Annu. Rev. Immunol. 23, 225–274 (2005).
Kovacic, M. B. et al. Variation of the killer cell immunoglobulin-like receptors and HLA-C genes in nasopharyngeal carcinoma. Cancer Epidemiol. Prev. Biomarkers 14, 2673–2677 (2005).
Vilches, C. & Parham, P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu. Rev. Immunol. 20, 217–251 (2002).
Andre, P. et al. New nomenclature for MHC receptors. Nat. Immunol. 2, 661–661 (2001).
Faure, M. & Long, E. O. KIR2DL4 (CD158d), an NK cell-activating receptor with inhibitory potential. J. Immunol. 168, 6208–6214 (2002).
Marsh, S. G. et al. Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Immunogenetics 55, 220–226 (2003).
Hsu, K. C., Chida, S., Geraghty, D. E. & Dupont, B. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunol. Rev. 190, 40–52 (2002).
Li, H., Wright, P. W., McCullen, M. & Anderson, S. K. Characterization of KIR intermediate promoters reveals four promoter types associated with distinct expression patterns of KIR subtypes. Genes Immun. 17, 66 (2016).
Wilson, M. J. et al. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc. Natl. Acad. Sci. 97, 4778–4783 (2000).
Gómez-Lozano, N., Gardiner, C. M., Parham, P. & Vilches, C. Some human KIR haplotypes contain two KIR2DL5 genes: KIR2DL5A and KIR2DL5B. Immunogenetics 54, 314–319 (2002).
Pyo, C.-W. et al. Different patterns of evolution in the centromeric and telomeric regions of group A and B haplotypes of the human killer cell Ig-like receptor locus. PLoS ONE 5, e15115 (2010).
Middleton, D. & Gonzelez, F. The extensive polymorphism of KIR genes. Immunology 129, 8–19 (2010).
Martin, A. M. et al. Comparative genomic analysis, diversity and evolution of two KIR haplotypes A and B. Gene 335, 121–131 (2004).
González-Galarza, F. F. et al. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 43, D784–D788 (2014).
Yin, C. et al. Genetic polymorphism and evolutionary differentiation of Eastern Chinese Han: a comprehensive and comparative analysis on KIRs. Sci. Rep. 7, 1–13 (2017).
Hernandez, E. G. et al. Genotype B of killer cell immunoglobulin-like receptor is related with gastric cancer lesions. Sci. Rep. 8, 6104 (2018).
Vierra-Green, C. et al. Allele-level haplotype frequencies and pairwise linkage disequilibrium for 14 KIR loci in 506 European-American individuals. PLoS ONE 7, e47491 (2012).
Nemat-Gorgani, N. et al. Diversity of KIR, HLA class I, and their interactions in seven populations of Sub-Saharan Africans. J. Immunol. 202, 2636–2647 (2019).
Jiang, W. et al. Copy number variation leads to considerable diversity for B but not A haplotypes of the human KIR genes encoding NK cell receptors. Genome Res. 22, 1845–1854 (2012).
Cisneros, E., Moraru, M., Gómez-Lozano, N., López-Botet, M. & Vilches, C. KIR2DL5: an orphan inhibitory receptor displaying complex patterns of polymorphism and expression. Front. Immunol. 3, 289 (2012).
Hou, L., Chen, M., Ng, J. & Hurley, C. Conserved KIR allele-level haplotypes are altered by microvariation in individuals with European ancestry. Genes Immun. 13, 47–58 (2012).
Cooley, S. et al. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood 116, 2411–2419 (2010).
Ashouri, E., Farjadian, S., Reed, E. F., Ghaderi, A. & Rajalingam, R. KIR gene content diversity in four Iranian populations. Immunogenetics 61, 483–492 (2009).
Uhrberg, M. et al. Human diversity in killer cell inhibitory receptor genes. Immunity 7, 753–763 (1997).
Carrington, M. et al. Hierarchy of resistance to cervical neoplasia mediated by combinations of killer immunoglobulin-like receptor and human leukocyte antigen loci. J. Exp. Med. 201, 1069–1075 (2005).
Omosun, Y. O. et al. Association of maternal KIR gene content polymorphisms with reduction in perinatal transmission of HIV-1. PloS One 13 (2018).
Canossi, A. et al. Role of KIR and CD16A genotypes in colorectal carcinoma genetic risk and clinical stage. J. Transl. Med. 14, 239 (2016).
Burkitt, D. P. Etiology of Burkitt’s lymphoma—an alternative hypothesis to a vectored virus. J. Natl Cancer Inst. 42, 19–28 (1969).
Zhou, Q. et al. KIR diversity in three ethnic minority populations in China. J. Transl. Med. 13, 1–8 (2015).
Nakimuli, A. et al. Killer cell immunoglobulin-like receptor (KIR) genes and their HLA-C ligands in a Ugandan population. Immunogenetics 65, 765–775 (2013).
Ozturk, O. G., Polat, G. & Atik, U. Diversity of killer cell immunoglobulin-like receptor genes in Southern Turkey. Mol. Biol. Rep. 39, 1989–1995 (2012).
Alicata, C. et al. KIR variation in Iranians combines high haplotype and allotype diversity with an abundance of functional inhibitory receptors. Front. Immunol. 11, 556 (2020).
Williams, F. & Middleton, D. KIR allele frequencies in a Xhosa population from South Africa. Hum. Immunol. 9, 1084–1085 (2004).
Yindom, L. M. et al. Killer-cell immunoglobulin-like receptors and malaria caused by Plasmodium falciparum in The Gambia. Tissue Antigens 79, 104–113 (2012).
Tukwasibwe, S. et al. Diversity of KIR genes and their HLA-C ligands in Ugandan populations with historically varied malaria transmission intensity. Malar. J. 20, 1–11 (2021).
Barani, S., Hosseini, S. V. & Ghaderi, A. Activating and inhibitory killer cell immunoglobulin like receptors (KIR) genes are involved in an increased susceptibility to colorectal adenocarcinoma and protection against invasion and metastasis. Immunobiology 224, 681–686 (2019).
Buckle, G. et al. Factors influencing survival among Kenyan children diagnosed with endemic Burkitt lymphoma between 2003 and 2011: a historical cohort study. Int. J. Cancer 139, 1231–1240 (2016).
Piriou, E. et al. Early age at time of primary Epstein-Barr Virus infection results in poorly controlled viral infection in infants from Western Kenya: clues to the Etiology of Endemic Burkitt Lymphoma. J. Infect. Dis. 205, 906–913 (2012).
Moormann, A. M. et al. Exposure to holoendemic malaria results in elevated Epstein-Barr virus loads in children. J. Infect. Dis. 191, 1233–1238 (2005).
Münz, C. & Chijioke, O. Natural killer cells in herpesvirus infections. Research 6, 1231 (2017).
Azzi, T. et al. Role for early-differentiated natural killer cells in infectious mononucleosis. Blood 124, 2533–2543 (2014).
Orange, J. S. Natural killer cell deficiency. J. Allergy Clin. Immunol. 132, 515–525 (2013).
Movassagh, M., Oduor, C., Forconi, C., Moormann, A. M. & Bailey, J. A. Sensitive detection of EBV microRNAs across cancer spectrum reveals association with decreased survival in adult acute myelocytic leukemia. Sci. Rep. 9, 1–10 (2019).
Omar, S. Y. A. et al. Genotypic diversity of the Killer Cell Immunoglobulin-like Receptors (KIR) and their HLA class I Ligands in a Saudi population. Genet. Mol. Biol. 39, 14–23 (2016).
Kimura, H. et al. Quantitative analysis of Epstein-Barr virus load by using a real-time PCR assay. J. Clin. Microbiol. 37, 132–136 (1999).
Acknowledgements
We thank the Kenyan children who participated in this study and Priya Saikumar-Lakshmi for participating in performing the assays. BMM was supported by the US National Institute of Health, National Cancer Institute R01 CA189806 (AMM), and DELTAS Africa grant (DEL-15-007: Awandare). The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)'s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa's Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust (107755/Z/15/Z: Awandare) and the UK government. We also wish to thank the Director of the Kenya Medical Research Institute (KEMRI) for approving this manuscript for publication. The views expressed in this publication are those of the author(s) and not those of the funders.
Author information
Authors and Affiliations
Contributions
B.M.M. conceived, participated in the design, performed KIR genotyping, performed statistical analyses, prepared and edited the manuscript. J.M.O., A.M.M., J.A.B., and A.G. conceived, designed and coordinated assay performance, prepared, and edited the manuscript. C.S.F. and P.O.O. edited the manuscript and participated in statistical analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Muriuki, B.M., Forconi, C.S., Oluoch, P.O. et al. Association of killer cell immunoglobulin-like receptors with endemic Burkitt lymphoma in Kenyan children. Sci Rep 11, 11343 (2021). https://doi.org/10.1038/s41598-021-90596-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-90596-7
This article is cited by
-
Compromised activity of natural killer cells in diffuse large b-cell lymphoma is related to lymphoma-induced modification of their surface receptor expression
Cancer Immunology, Immunotherapy (2023)
-
The impact of HLA polymorphism on herpesvirus infection and disease
Immunogenetics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.