Abstract
While it is now broadly accepted that inter-individual variation in the outcomes of host–pathogen interactions is at least partially genetically controlled, host immunogenetic characteristics are rarely investigated in wildlife epidemiological studies. Furthermore, most immunogenetic studies in the wild focused solely on the major histocompatibility complex (MHC) diversity despite it accounts for only a fraction of the genetic variation in pathogen resistance. Here, we investigated immunogenetic diversity of the Alpine ibex (Capra ibex) population of the Bargy massif, reservoir of a virulent outbreak of brucellosis. We analysed the polymorphism and associations with disease resistance of the MHC Class II Drb gene and several non-MHC genes (Toll-like receptor genes, Slc11A1) involved in the innate immune response to Brucella in domestic ungulates. We found a very low neutral genetic diversity and a unique MHC Drb haplotype in this population founded few decades ago from a small number of individuals. By contrast, other immunity-related genes have maintained polymorphism and some showed significant associations with the brucellosis infection status hence suggesting a predominant role of pathogen-mediated selection in their recent evolutionary trajectory. Our results highlight the need to monitor immunogenetic variation in wildlife epidemiological studies and to look beyond the MHC.
Similar content being viewed by others
Introduction
The epidemiological dynamics of infectious diseases results from a complex interplay between the pathogen, the host and the environment1. Often neglected, inter-individual variation in host susceptibility plays a key role in this dynamics since, depending on their biology (e.g. age, sex), their spatial or social behaviours and/or their genetic characteristics, some individuals contribute far more than others to disease spread2. Therefore, identifying individuals and their characteristics that are most responsible for disease transmission is an important step for improving epidemiological surveillance and management of wildlife diseases3.
Host genetic and immunogenetic characteristics are rarely investigated in wildlife epidemiological studies despite evidence that inter-individual variation in the outcomes of host–pathogen interactions is at least partially genetically controlled4. Pathogen-mediated selective pressures shape the genetic components of host immunity and give rise to inter-individual variation in resistance to infectious diseases5. This immunogenetic variation is particularly well documented in humans and domestic animals but much less in wild animals. Furthermore, most studies focused solely on the adaptive branch of the immune system, particularly on the major histocompatibility complex (MHC)6,7. Although there is no doubt that MHC plays a key role in individual susceptibility to many diseases8, it accounts for only a fraction of the genetic variation in pathogen resistance5,9. Before the adaptive immunity intervenes in the host response, other genes involved in the innate immunity such as the Toll-like receptors (TLRs) genes are major players in the host frontline defense against a wide range of microparasites including vectors of zoonotic diseases such as Coxiella brunetii (Q fever) or Borrelia sp. (Lyme disease)10,11,12.
Genetic variation at the MHC and other immunity-related genes encoding pathogen recognition receptors is shaped by both neutral (i.e. genetic drift, mutation, migration) and selective processes. The relative importance of these evolutionary forces depends on both the population demographic history that influences the strength of the genetic drift, and the (multi)pathogen challenge that determines the type and intensity of selection processes13. Populations that went through severe bottlenecks may show low levels of genetic diversity at the MHC (Mainguy et al.14 on the mountain goat Oreamnos americanus; Bollmer et al.15 on Galapagos penguin Spheniscus mendiculus) or TLR loci (Grueber et al. 2013 on Stewart Island robin Petroica australis rakiura). However pathogen-mediated balancing selection may in some cases buffer the effect of drift to maintain polymorphism16,17. Several not-mutually exclusive mechanisms may act in synergy to maintain immunity-related diversity, the three more widespread being: the “negative frequency-dependence”, the “heterozygous advantage” and the “fluctuating selection”. The “negative frequency-dependent selection” (also called rare-allele advantage) hypothesis suggests that the fitness-advantage of alleles is inversely proportional to their frequency in the population as a result of the host-pathogen18 or prey-predator co-evolutionary dynamics19. The “heterozygote advantage” hypothesis predicts a correlation between individual heterozygosity and fitness or disease susceptibility20,21. The “fluctuating selection” hypothesis22,23 suggest that population genetic diversity is maintained when spatially and/or temporally varying directional selection occurs24.
Here, we investigated the neutral and immunity-related genetic diversity of the Alpine ibex (Capra ibex) population from the Bargy area in France. This ibex population from the northern French Alps has undergone a major outbreak of brucellosis (Brucella melitensis). Until very recently, European wild ungulates were seen as dead-end hosts for this pathogen25 and all previous outbreaks of brucellosis in wild ruminants spontaneously faded out26,27,28. However, the situation in Bargy is alarming since the disease has affected an unprecedented high proportion of the population (38% in 2013, 95% CI [28.2; 47.8]; n = 77). Brucellosis persisted at high levels in the following years despite intense culling of animals which led to the removal of 40% of the population (see Marchand et al.29 for details). Beyond the conservation and ethical issues, this outbreak has raised public health and economic concerns since, for the first time, cattle and humans were infected by the wildlife reservoir30. In this context, it was critical to identify the drivers of pathogen persistence in this population and individual characteristics favoring its spread. Previous works showed that host-infectiousness vary significantly across age and sex and among socio-spatial units within the Bargy massif29. However, genetic factors of susceptibility to brucellosis in Alpine ibex have never been investigated while several associations with innate gene polymorphism have been discovered in domestic ungulates31,32.
The genetic issue is particularly pertinent in Alpine ibex since this species underwent a major bottleneck during the XIXth century due to overhunting, followed by several successive bottlenecks during its reintroduction history across the Alpine arc. Most current populations have been re-established few decades ago from a limited number of founder individuals33. This reintroduction history has had drastic genetic consequences: all Alpine ibex populations exhibit a particularly low level of neutral and MHC genetic diversity and a high level of inbreeding33,34 with a direct influence on traits related to survival and reproductive success such as body mass or parasite load35.
The first aim of this study was to assess the neutral and immunity-related genetic variation of the Bargy population. We screened the neutral microsatellite diversity to test the hypothesis that the intrinsic susceptibility to brucellosis of ibex at Bargy results from a particular low genetic diversity in comparison with other reintroduced ibex populations. We also evaluated the polymorphism of the commonly used Mhc-Drb as well as four immunity-related genes (Tlr1, Trl2, Tlr4, Solute Carrier family 1 hereafter SLC11A1) involved in the innate immune response to Brucella sp. in domestic ungulates31,32,36. The second objective was to investigate associations between individual brucellosis infection status and genetic/immunogenetic profile. We first assessed the role of multi-locus heterozygosity at neutral microsatellites as a proxy of inbreeding (heterozygosity-fitness correlation). We then looked at potential associations with the heterozygosity status (heterozygous advantage hypothesis) and/or specific haplotype of immunity-related genes. In both domestic and wild ungulates, brucellosis is well-known to cause abortion and reproductive failures37,38 with known impacts on individual fitness. Therefore, these associations may reflect pathogen-mediated balancing or directional selective pressure on immunity-related gene polymorphism.
Results
Neutral genetic diversity
The tests for Hardy-Weinberg equilibrium, after correction for multiple testing, revealed that SR-CRSP07 STR locus showed a significant excess of homozygotes. This marker was thus removed from further analysis. HE was 0.43 ± 0.17 (mean ± standard error) and mean NA was 3.12 over all microsatellite loci. Neutral variation did not vary significantly before (HE = 0.42 ± 0.16, sNA = 2.64 ± 0.79) and after (HE = 0.42 ± 0.17, NA = 2.78 ± 0.84) the culling operations that occurred in 2013–2015 (Student’s t-test, df=109, p = 0.93 for HE; p = 0.39 for sNA).
Immunity-related gene polymorphism
The Illumina sequencing of the exon 2 of the Mhc-Drb gene on 144 individuals revealed a unique haplotype in Bargy corresponding to CaIb-DRB*1 haplotype, previously isolated by Schalschl et al.39 and Grossen et al.40. The Sanger sequencing of the three Tlr genes generated an average of 2,169 bp of sequence covering almost the entire coding region of the genes (92% on average). Tlr genes exhibited seven SNPS among which five were non-synonymous. We isolated 1 SNPs for Tlr1 and revealed two functional haplotypes (i.e. encoding different amino acid sequences) including a rare haplotype (5%) observed in only 14 individuals. The Tlr2 gene exhibited three SNPs (two non-synonymous) and showed three functional haplotypes with a common haplotype (Tlr2a = 72%) and two haplotypes with low frequencies (Tlr2b = 15% and Tlr2c = 13%). Lastly, we found three SNPs (two non-synonymous) in Tlr4 gene and identified four functional haplotypes among which three haplotypes showed similar moderate frequencies ranging between 25% and 35% while the fourth one was rare (9%). Expected heterozygosity (HE) (given differences in haplotype frequencies) varied greatly among Tlrs ranging from 0.09 for Tlr1 to 0.71 for Tlr4 (Table 1). The SLC11A1 gene showed two alleles (“A324” and “A330” hereafter) with frequencies of 75% and 25% respectively (HE = 0.37).
Genetic effects on brucellosis infection
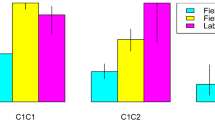
Out of the 237 ibex from the Bargy massif that were first tested, 89 were found seropositive to brucellosis (37%). The probability to be seropositive was lower in males than in females and reached a maximum in the core area of the massif (SSU3, SSU4) (see Tables S4,S5, Figs. 1,2). We observed a slight temporal decrease of the seroprevalence over the monitoring period (only significant for the dataset covering the whole 2012–2017 study period). Brucellosis seropositivity also tended to increase with age but 95% confidence intervals of the effect size did not exclude zero (Average Effect Size = 0.41 [95% CI: −0.13,0.95]) (Fig. 1, Table S4). We did not find any significant effect of the multi-locus heterozygosity (MLH) on the individual serological status (general effect) (Fig. 2, Table S5) but we observed associations with immunity-related genes. First, we found a strong relationship between SLC11A1 genotype and brucellosis status (Fig. 1). Prevalence decreased with the number of copies of the less frequent allele “A330”. This effect was particularly marked for individuals carrying two copies of this allele (homozygous individuals) (AES = −2.36 [−4.60, −0.13]) but less clear for heterozygous individuals (AES = −0.24 [−0.86, 0.39]), which suggests that “A330” is recessive. We also revealed a significant association with Tlr1 genotype (Fig. 2, Table S5): homozygous individuals carrying two copies of the frequent haplotype (Tlr1a) had a lower probability to be seropositive than heterozygous individuals (AES = −1.90 [−3.52, −0.29]). Tlr1 heterozygous status was also associated with ibex brucellosis status (Table S5) but this most likely result from the systematic presence of the Tlr1b haplotype in Tlr1 heterozygous ibex. We did not find any effect of Tlr2 and Tlr4 haplotypes (directional selection) or of the heterozygous/homozygous status (heterozygote advantage) (Table S5).
Genetic association between Slc11A1 and brucellosis serological status. Model averaged parameter estimates and their 95% confidence intervals for the Slc11A1 genotype, biological (Age, Sex) and environmental factors (Year, Socio-spatial units). The symbol * indicates a parameter with a significant effect. Females from SSU2 sampled in 2012 were used as the reference category.
Genetic effects of neutral multi-locus heterozygosity (MLH) and Tlr polymorphism on brucellosis serological status. Model averaged parameter estimates and their 95% confidence intervals for the MLH (Multi-locus heterozygosity estimated from neutral microsatellite loci), Tlr (Tlr1, Tlr2, Tlr4) number of copies (0, 1 or 2) of each haplotype with frequency >10%, Tlr heterozygosity status (Het), biological (Age, Sex), and environmental factors (Year, Socio-spatial units). The symbol * indicates a parameter with a significant effect. Females from SSU2 sampled in 2012 were used as the reference category.
Discussion
The aim of this study was to assess the neutral and immunity-related genetic diversity of the Alpine ibex population of the Bargy massif, wildlife reservoir of a persistent and virulent outbreak of brucellosis. As expected, this population established a few decades ago from a very limited number of founders (<15 ibex) shows very low genetic variation at neutral microsatellite loci and exhibits a single MHC Class II haplotype similarly to most ibex populations of the Western Alps40. By contrast, other immunity-related genes involved in the innate immunity such as Tlr genes (Tlr1, Tlr2 and Tlr4) and Slc11A1 have maintained polymorphism despite serial founder events occurring throughout the Alpine ibex reintroduction history. We revealed significant associations between the polymorphism of some of these genes (Tlr1 and Slc11A1) and Brucella infection status, which may suggest a role of pathogen-mediated selection in their recent evolution.
Low neutral genetic diversity and absence of genome-wide effect on brucellosis infection
With an expected heterozygosity (HE) of ~40% and an average number of alleles (MNA) per microsatellite loci <4, the ibex population of Bargy shows particularly low levels of microsatellite genetic variation, compared to values reported in goat breeds (MNA = 5–9 alleles, HE = 0.60–0.76, Canon et al. 2006) and in other wild European ungulate populations (e.g. HE = 0.56–0.67 in roe deer Capreolus capreolus, Quéméré et al.17; HE = 0.58–0.67 in red deer Cervus elaphus, Zachos et al.41). However, these differences may arise because the microsatellite loci used in the different species underwent different ascertainment bias. Our results are consistent with the levels previously observed in other Alpine ibex populations reintroduced in Switzerland during the XXth century42 and in particular, for the Mont Pleureur population (HE = 0.42) from which the Bargy founders came from. Low genetic diversity might reduce individual fitness and population demographic performance due to reduced evolutionary potential43,44. For example, in the Gran Paradiso ibex population, there is a direct relationship between multi-locus microsatellite heterozygosity, male body mass and gastrointestinal nematode infestation (fecal egg counts) (i.e. Heterozygosity-fitness correlation)35. In our case, we did not observe any relationship between the genome-wide heterozygosity (MLH) of ibex and their Brucella infection status. This is in agreement with the recent study of Brambilla et al.45 that did not find any effect of neutral heterozygosity on infectious keratoconjunctivitis symptoms, another bacterial disease recurrently affecting Alpine ibex populations. Similar low levels of neutral genetic diversity were observed in other brucellosis-infected ibex populations (Grand Paradiso) in which the prevalence level has remained low and infection vanished spontaneously26. Therefore, contrary to our hypothesis, the low level of neutral diversity at Bargy is in itself not sufficient to explain its potential higher sensitivity to brucellosis compared to other ibex populations. However, a growing number of studies illustrated the poor predictive power of microsatellite loci to evaluate functional diversity and showed the importance of accounting for immunogenetic variation to get a better understanding of the population’s adaptive response potential to pathogen-mediated selection46,47.
Maintenance of immunity-related gene polymorphism in Alpine ibex
Our study is the first to our knowledge to reveal immunity-related polymorphism in non-MHC genes in Alpine ibex. Previous work on ibex focused exclusively on MHC Class II genes45,48 that accounts for only a fraction of the genetic variation in pathogen resistance5,9. MHC class II molecules principally bind exogenous antigens and are primarily involved in the immune response to extracellular pathogens49. By contrast, genes encoding toll-like receptors (Tlrs) play a fundamental role in vertebrate innate immune defense against intracellular and extracellular micropathogens including viruses, bacteria, protozoa or fungi50. In particular, these genes are involved in the recognition and immune response to pathogenic bacteria that recurrently cause outbreaks such as Mycoplasma sp. (causing pneumonia or infectious kerotoconjunctivitis infections)51,52 or Brucella abortus53,54. Contrary to our expectation and despite serial founder events throughout the species reintroduction history, the three studied Tlr genes and the SlC11A1 have all maintained several haplotypes while the MHC Class II DRB (second exon) is monomorphic in Bargy. There are very few studies that investigate Tlr polymorphism in ungulates populations: levels of Tlr haplotype diversity in Bargy ibex (NH = 3–5, HD) appears similar to those reported for the same genes in roe deer populations (NH = 3–5)17, domestic goat (NH = 1–4)55 or cattle (NH = 2–5)56. Tlr4 showed four functional haplotypes (i.e. encoding different amino-acid sequences) including three alleles with moderate to high frequencies (>25%). This balanced polymorphism which contrasts with the very low neutral and MHC diversity in Bargy, suggests that balancing pathogen-mediated selection may have favoured the maintenance of genetic variation as showed in other free-ranging animal species12,57,58. Furthermore, it is worth noting that the presence of balanced polymorphism does not mean that the gene is involved in brucellosis resistance, which can be demonstrated only by studying associations between immunogenetic patterns and pathogen prevalence59.
Genetic factors of resistance to Brucella melitensis infection in Alpine ibex
In contrast to Brambilla et al.35, we did not find evidence of correlation between multi-locus heterozygosity at neutral microsatellites and brucellosis infection status (“general effect” hypothesis). This means that a higher individual multi-locus heterozygosity (a proxy of the level of inbreeding) does not confer a resistance advantage regarding Brucella infection. In contrast, we found associations between single immunity-related genes (here Tlr1, Slc11A1) and ibex infection status hence supporting the “local effect” hypothesis in agreement with Brambilla et al.45. These authors observed a lower susceptibility of MHC Drb heterozygous to infectious keratoconjunctivitis (heterozygote advantage hypothesis) while we revealed here an effect of specific alleles. Ibex carrying two copies of the less frequent allele of the Slc11A1 gene were less likely to develop the Brucella infection, which suggests that this “resistance allele” is recessive. Associations between a specific Slc11A1 allele and Brucella infection has been already reported in domestic goat36 and water buffalo31. Moreover, in an in-vitro experiment on water buffalo’s monocytes infected by several Brucella species including B. melitensis, Borriello et al.60 demonstrated that some Slc11A1 genetic variants confer a higher Slc11A1 mRNA expression and a higher ability in controlling the intracellular replication of the Brucella. The “resistant allele” (A330) showed similar frequency in the Bargy and Aravis populations (25% and 21% respectively) but is much more frequent in other Alpine populations (e.g. 54% in Belledonne massif (northern French Alps), 40% in Grand Paradiso National Park – results not shown). Further comparison with other recently reintroduced ibex populations should be performed to investigate whether the particularly low frequency of this resistant variant in Bargy may have favored the emergence of this brucellosis outbreak together with other, non-genetic factors such as spatial ibex distribution and abundance.
Our results also suggest that the most frequent haplotype of Tlr1 (96%) conferred a resistance advantage against Brucella infection. However, this resistance advantage is relative: although 92% of ibex in Bargy are homozygous for this allele, 37% are still seropositive. Therefore, individuals that carry out this allele have a lower probability to get infected after a contact with the bacteria, but do not show a complete resistance. Such relationship between Tlr1 genotype and Brucella infection has been already evidence in Cattle by Prakash et al.32. Tlr1 interact with Brucella sp. by recognizing specific pathogen-associated molecular patterns. Individuals carrying the rare allele (Tlr1b) may have a decreased ability to recognize Brucella and thus to trigger an adequate immune response. Further work is needed to clarify the functional role of these immunity-related genes against brucellosis in Alpine ibex and in particular how these polymorphisms affects the detection and the replication of the bacteria (see Borriello et al.60). Particularly, in vitro studies of ibex white blood cells and their response to Brucella36 would help to confirm a variation linked to immune genes and to identity cellular pathways to resistance.
In agreement with our hypothesis and the literature on domestic ungulates, we showed significant associations between immunity-related genes (specific alleles Slc11A1 and Tlr1) and brucellosis infection status in Alpine ibex. However, this does not in itself provide evidence for directional pathogen-mediated selection because we did not actually demonstrate that these alleles confer a selective advantage. Seropositive ibex were euthanized, so there is no data available on the impact of B. melitensis infection on survival and reproduction rates in this population. However, brucellosis is well-known to cause abortion and reproductive failures in other domestic and wild ungulates37,38. So we can reasonably hypothesize that B. melitensis infection actually affects the fitness of ibex in Bargy and may ultimately lead to selection on these genes.
Management implications
Genetic monitoring is widely recognized as a valuable tool for the management and conservation of populations61. In particular, access to population genetic parameters may help to better understand the ecology of host/pathogen interaction and thus better manage wildlife diseases62,63. Most genetic studies used microsatellites and MHC Class II markers to investigate population adaptive diversity and tolerance or resistance to diseases8 with very few exceptions64. However, Alpine ibex exhibit a very low neutral genetic diversity and a unique MHC Class II DRB exon 2 haplotype in most recently introduced populations. Furthermore, most potentially zoonotic diseases affecting wild ungulate populations and notably Alpine ibex (e.g. Q fever, tuberculosis, chlamydiosis) involve micropathogens (virus, bacteria, protozoa) and MHC Class II often plays a minor role in the immune response against these pathogens49. Our study highlights the need to look beyond MHC class II and to explore the diversity of other candidate immunity-related genes5, in particular MHC class I, cytokine and Toll-like receptors genes that are centrally involved in the innate immune response against micro pathogens in humans and domestic species.
In a conservation framework, our results call for immediate management actions to increase the neutral and immunity-related diversity of the Bargy ibex population, whose census size had been halved (~300 individuals in 2014–2017 against ~600 in 2013) over the monitoring period. Translocations of unrelated individuals from the Gran Paradiso source population or from other genetic units of Alpine ibex34,65 is recommended to prevent inbreeding depression and increase both genome-wide35,45 and immunity-related diversity47. Further, these results raise the question of the interplay between sanitary management and population genetic structure. Here, the test-and-cull management procedure was expected to affect the genetic structure of the population, by selectively eliminating infected individuals carrying specific alleles. In the same line, some authors suggested that artificial selection of specific Slc11A1 or Tlr genotypes in breeding programs may help to increase population natural resistance and limit spread of brucellosis in domestic ungulates31,32. However, it is important to keep in mind that natural populations are exposed to multi-pathogen pressures (e.g. brucellosis and Q fever in the Bargy population) and that most immunity-related genes are involved in the immune response against several pathogens. Therefore, artificially selecting a specific genotype to increase population resistance to a pathogen may lead to the loss of key haplotypes in the host-control of other diseases, particularly in species with very low genetic diversity. Incorporating data on immunogenetic variation in epidemiological models should help to predict how variation in allele frequency may affect the spread dynamics of the disease, and how disease management may affect genetic structure of the population.
Methods
Study site, population and sampling
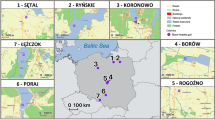
Alpine ibex were reintroduced in the Bargy Massif (France) during three release events in 1974 and 1976 comprising a total of 14 released individuals (i.e. 6 males and 8 females) translocated from the Mont Pleureur population in Switzerland66. Since the discovery of the outbreak in 2012, the population has been continuously monitored by Capture-Mark-Recapture (CMR)29. Using GPS data on marked individuals, Marchand et al.29 demonstrated that females are structured in five distinct socio-spatial units (SSU1-SSU5, Fig. 3), whereas males are prone to move between units especially during the mating period29. Here, we studied the genetic and immunogenetic characteristics of 262 ibex captured between 2012 and 2017 by the French Hunting and Wildlife Agency in accordance with legal and ethical regulations (French environmental code, 2005, 2006; Préfecture de Paris, 2009; Préfecture de la Haute-Savoie, 2013, 2015a,b; French Minister of Ecology Sustainable Development and Energy, 2014). Animals were captured by dart-gun xylazine-ketamine anesthesia (Rompun®, Bayer, Leverkusen, Germany and Imalgène®, Merial, Lyon, France; 100 mg/individual) following protocols described in Lambert et al.67. During captures, test-and-cull was implemented on the basis of serological tests (see below for further details on the serological method) as part of management measures decided by the French Authorities (Hars et al., 2013). Seronegative individuals were marked and released while seropositive ones were euthanized. All captured animals were handled by professionals from the French Game and Wildlife management agency (Office National de la Chasse et de la Faune Sauvage, Office Français de la Biodiversité) according to the ethical conditions detailed in the specific accreditations delivered by the Préfecture de Paris (prefectural decree 2009–014), by the French Minister of Ecology, Sustainable Development and Energy (Ministerial Orders of February 11, 2014) and by the Préfecture de la Haute-Savoie (prefectural decree 2015062- 0018) in accordance with the French environmental code (Art. R421-15 to 421-31 and R422-92 to 422-94-1). Euthanasia was performed by veterinarians in accordance with the requirements of these accrediting authorities, and slaughtering operations were performed in accordance with accreditations delivered by the Préfecture de la Haute-Savoie (prefectural decrees 2013274-0001, DDT-2015-0513, DDT-2016-0933, DDT-2017-1003). We hence confirm that we treated animals in accordance with relevant guidelines and regulations and that our protocols were approved by the National Committee for Nature Protection (CNPN in French), a group of independent experts elicited by the ministry of the environment and in charge of evaluating capture/destruction of the protected species.
Location of the five socio-spatial units (SSU) of female Alpine ibex. These five socio-spatial units correspond to the best number of spatially-segregated groups as determined by hierarchical classification methods on distances between individuals measured as overlap between annual home ranges of GPS-collared females. Adapted from Marchand et al.29 (CC by 4.0 International license).
In addition to CMR monitoring, French Authorities implemented selective culling of animals with observed clinical signs and mass indiscriminate slaughter of individuals of five years and more: culling operations occurred between autumn 2013 and spring 2014 (N = 256 killed) and then in autumn 2015 (N = 70). The population size dropped from 567 individuals (95% CI: [487–660]) in 2013 to 310 [275–352] in 2014 and 277 [220–351] in 201529.
Pathogen screening
Brucellosis serologic status of individuals was evaluated using a range of serological tests68. In particular, since 2014, the rapid Laminar Flow Immune-chromatographic Assay (LFIA) (Rapid G.S. Brucella Abtest, Bionote, Gyeonggi-do, Rep. of Korea) was performed in the field. This test had been validated on ibex samples by the EU/OIE/FAO and National reference laboratory (ANSES, 2014). Further details on the validation method and the congruence of the FLIA with reference tests (Rose Bengal Test, Complement Fixation Test, iELISA) were provided in Anses (2014). Seropositive ibex were considered as infected by B. melitensis, but not necessarily infectious at the time of capture67.
Microsatellite and immunity-related loci genotyping
DNA was extracted from blood using the DNeasy Blood and Tissue kit (QIAGEN). All individuals were genotyped at 25 polymorphic putatively neutral microsatellites42 (see list and primers in Table S1). We carried out genotyping replicates for about 25% of the samples to ensure the reliability of the genotyping. Micro-Checker 2.2.369 was employed to assess the frequency of null alleles and scoring errors (stuttering or large allelic dropout). The same individuals were also typed at a microsatellite located at the 3′UTR of the Solute Carrier Family 11 member A1 (SLC11A1) gene using the following primers: Fw- TCTGGACCTGTCTCATCACC and Rv- ACTCCCTCTCCATCTTGCTG70. The SLC11A1 gene, formerly known as natural resistance-associated macrophage protein 1 (NRAMP1) gene, is involved in the innate resistance to intra-cellular pathogens71. Polymorphism in microsatellites of the 3′ UTR of the gene were associated with resistance to Brucella infection in water buffalo Bubalus bubalis36 and in the domestic goat31, a species closely related to the Alpine ibex. In particular, some SLC11A1 genotypes have a higher ability of controlling the intracellular replication of Brucella in vitro60.
We also genotyped Mhc-Drb and Tlr genes for a subset of the individuals (N = 146) sampled between 2012 and 2014. The second exon of the Mhc-Drb class II gene encoding the ligand-binding domain of the protein was amplified and sequenced using Illumina MiSeq system following the procedure detailed in Quéméré et al.17. Haplotypes and individual genotypes were identified using the SESAME barcode software72. Tlr genes (Tlr1, Tlr2 and Tlr4) were genotyped using the two-step procedure described in Quéméré et al.17. A pilot study on 30 individuals was first performed to identify polymorphic sites (SNPs). We screened almost the entire coding region of the three Tlr genes (92% in average) including the leucine-rich extracellular region of receptors involved in antigen-binding. Details on primer sequences and accession numbers are provided in Table S2. SNPs were then genotyped for 146 individuals using the KASPar allele-specific genotyping system provided by KBiosciences (Hoddesdon, UK). Details on SNP position and codon change can be found in Table S3. Haplotypes were then reconstructed from the phased SNPs using the procedure implemented in DNASP v573.
Genetic diversity
The genetic diversity of both microsatellites and immunity-related genes was evaluated by calculating the expected heterozygosity (HE) and the number of alleles/haplotypes (NA) as implemented in GENETIX software v4.05.274. For each locus, a test for Hardy–Weinberg proportions was performed based on 1000 permutations. To allow comparison with other ibex populations42 and between the two monitoring periods (before and after culling operations), we used FSTAT version 2.9.375 to calculate the allelic richness (sNA), a standardized measure of number of alleles corrected for differences in samples size. Lastly, standardized multi-locus heterozygosity at neutral microsatellites (MLH) was calculated for each individual as the ratio of its heterozygosity to the mean heterozygosity of the population76. Our set of microsatellite loci showed significant identity disequilibrium (g2 ± SD = 0.011 ± 0.0065, p = 0.007), (e.g. a measure of covariance in heterozygosity), a condition for detecting heterozygosity-fitness correlations. Both MLH and g2 were calculated using the R package inbreedR77. To assess whether management operations (i.e. removal of 40% of the population between 2013 and 2015) had led to a decrease of the genetic diversity, we performed a comparison (using Student’s t-tests) of the allelic richness (sNA) and expected heterozygosity (HE) of ibex sampled before (between autumn 2012 and autumn 2013) and after (years 2016–2017) the slaughtering operations that occurred between autumn 2013 and autumn 2015.
Genetic association with brucellosis infection
We analyzed the relationships between the serological status of ibex and neutral or immunity-related gene variation using generalized linear models (GLM) of the binomial family (using a logit link function). The serological status of each individual was coded as 1 (positive, considered to have been exposed and not resistant) or 0 (negative, considered to have been either non-exposed or exposed but resistant).
We fitted models testing for the effect of genetic effects, while taking into account other biological and environmental factors that possibly affect pathogen exposure. We tested the “heterozygosity fitness-correlation” hypothesis (i.e. “general effect”) by fitting individual standardized multi-locus microsatellite heterozygosity (MLH) as a fixed effect. For each immunity-related gene, we tested the “heterozygote advantage hypothesis” by fitting a binary fixed effect (heterozygote vs homozygote) and the “haplotype advantage” hypothesis by considering associations between the infection status and the number of copies (0, 1 or 2) of Tlr or Slc11A1 haplotypes with frequency >10%. Concerning non-genetic factors, following Marchand et al.29, we included the effects of ibex sex as a categorical variable and age as a continuous variable in years (linear and quadratic terms). We also fitted a “year” effect as a categorical variable to test for varying epidemiological situations across years and a “location” effect because seroprevalence has been shown to vary among socio-spatial units29.
We used a multimodel inference approaches to establish which explanatory variables had an effect, averaged over plausible models78,79,80. The most complete models used in the inference included the effects of genetic effects hypotheses, age sex, year and location. We compared the maximal model with all its sub-models (See Tables S6 and S7 for the full lists of model tested). The effect of Slc11A1 and Tlr genes were evaluated in separate analyses because of different samples sizes (262 and 146 individuals respectively). We conducted an initial exploration of our data to ascertain their distribution and spread, to identify outliers and examine relationships between variables81. We used variance inflation factors (VIF) to assess which explanatory variables were collinear, and should be retained in the analyses (i.e. VIF values <3)81. To minimize multicollinearity, the effects of the most frequent haplotype of a gene and its heterozygosity status were evaluated in separate models.
Model averaged parameter estimates are provided with their unconditional standard errors (SE) and 95% confidence intervals, after averaging model with ΔAIC < 7 (relative to the best model) as recommended by Burnham et al.79 and using the “zero method” (i.e. a zero is substituted into models where the parameter is absent). Analyses were performed using the package lme482, MUMIN v1.7.783 and AICcmodavg v1.2584 in R version 3.3.3 (R Development Core Team, 2017).
Data availability
Haplotype DNA sequences: primers and Genbank accessions are in Tables S1,S2. SNP positions and characteristics are in Table S3. Genotypes of all individuals at each microsatellite and immunity-related locus are available from the corresponding author upon request to anyone who wishes to repeat our analyses or collaborate with us.
References
Tompkins, D. M., Dunn, A. M., Smith, M. J. & Telfer, S. Wildlife diseases: from individuals to ecosystems. J. Anim. Ecol. 80, 19–38 (2011).
Galvani, A. P. & May, R. M. Epidemiology: dimensions of superspreading. Nat. 438, 293 (2005).
Matthews, L. et al. Heterogeneous shedding of Escherichia coli O157 in cattle and its implications for control. Proc. Natl Acad. Sci. 103, 547–552 (2006).
Wilfert, L. & Schmid-Hempel, P. The genetic architecture of susceptibility to parasites. BMC Evolut. Biol. 8, 187 (2008).
Acevedo-Whitehouse, K. & Cunningham, A. Is MHC enough for understanding wildlife immunogenetics? Trends Ecol. Evol. 21, 433–438 (2006).
Bernatchez, L. & Landry, C. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J. Evol. Biol. 16, 363–377 (2003).
Piertney, S. & Oliver, M. The evolutionary ecology of the major histocompatibility complex. Heredity 96, 7–21 (2005).
Sommer, S. The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Front. Zool. 2, 16 (2005).
Jepson, A. et al. Quantification of the relative contribution of major histocompatibility complex (MHC) and non-MHC genes to human immune responses to foreign antigens. Infect. Immun. 65, 872–876 (1997).
Ammerdorffer, A. et al. Recognition of Coxiella burnetii by Toll-like Receptors and Nucleotide-Binding Oligomerization Domain–like Receptors. J. Infect. Dis. 211, 978–987 (2014).
Netea, M. et al. Nucleotide-binding oligomerization domain-2 modulates specific TLR pathways for the induction of cytokine release. J. Immunol. 174, 6518–6523 (2005).
Tschirren, B. et al. Polymorphisms at the innate immune receptor TLR2 are associated with Borrelia infection in a wild rodent population. Proc. Biol. Sci. 280, 20130364 (2013).
Sutton, J. T., Nakagawa, S., Robertson, B. C. & Jamieson, I. G. Disentangling the roles of natural selection and genetic drift in shaping variation at MHC immunity genes. Mol. Ecol. 20, 4408–4420 (2011).
Mainguy, J., Worley, K., Côté, S. D. & Coltman, D. W. Low MHC DRB class II diversity in the mountain goat: past bottlenecks and possible role of pathogens and parasites. Conserv. Genet. 8, 885–891 (2006).
Bollmer, J. L., Vargas, F. H. & Parker, P. G. Low MHC variation in the endangered Galápagos penguin (Spheniscus mendiculus). Immunogenetics 59, 593–602 (2007).
Oliver, M. K. & Piertney, S. B. Selection maintains MHC diversity through a natural population bottleneck. Mol. Biol. evolution 29, 1713–1720 (2012).
Quéméré, E. et al. Immunogenetic heterogeneity in a widespread ungulate: the European roe deer (Capreolus capreolus). Molecular ecology (2015).
Takahata, N. & Nei, M. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genet. 124, 967–978 (1990).
Cook, L. M. The rise and fall of the carbonaria form of the peppered moth. Q. Rev. Biol. 78, 399–417 (2003).
Doherty, P. C. & Zinkernagel, R. M. Enhanced immunological surveillance in mice heterozygous at the H-2 gene complex. Nat. 256, 50 (1975).
Lewontin, R. C. & Hubby, J. L. A molecular approach to the study of genic heterozygosity in natural populations. II. Amount of variation and degree of heterozygosity in natural populations of Drosophila pseudoobscura. Genet. 54, 595 (1966).
Hill, A. V. HLA associations with malaria in Africa: some implications for MHC evolution. In Molecular evolution of the major histocompatibility complex 403–420 (Springer, 1991).
Levene, H. Genetic equilibrium when more than one ecological niche is available. Am. Naturalist 87, 331–333 (1953).
Wittmann, M. J., Bergland, A. O., Feldman, M. W., Schmidt, P. S. & Petrov, D. A. Seasonally fluctuating selection can maintain polymorphism at many loci via segregation lift. Proc. Natl Acad. Sci. 114, E9932–E9941 (2017).
Godfroid, J. et al. A “One Health” surveillance and control of brucellosis in developing countries: moving away from improvisation. Comp. immunology, microbiology Infect. Dis. 36, 241–248 (2013).
Ferroglio, E., Tolari, F., Bollo, E. & Bassano, B. Isolation of Brucella melitensis from alpine ibex. J. Wildl. Dis. 34, 400–402 (1998).
Garin-Bastuji, B., Oudar, J., Richard, Y. & Gastellu, J. Isolation of Brucella melitensis biovar 3 from a chamois (Rupicapra rupicapra) in the southern French Alps. J. Wildl. Dis. 26, 116–118 (1990).
Muñoz, P. M. et al. Spatial distribution and risk factors of brucellosis in Iberian wild ungulates. BMC Infect. Dis. 10, 46 (2010).
Marchand, P. et al. Sociospatial structure explains marked variation in brucellosis seroprevalence in an Alpine ibex population. Sci. Rep. 7, 15592 (2017).
Mailles, A. et al. Re-emergence of brucellosis in cattle in France and risk for human health. Eurosurveillance 17, 20227 (2012).
Iacoboni, P. A. et al. Polymorphisms at the 3′ untranslated region of SLC11A1 gene are associated with protection to Brucella infection in goats. Veterinary immunology immunopathology 160, 230–234 (2014).
Prakash, O. et al. Polymorphism of cytokine and innate immunity genes associated with bovine brucellosis in cattle. Mol. Biol. Rep. 41, 2815–2825 (2014).
Biebach, I. & Keller, L. F. Inbreeding in reintroduced populations: the effects of early reintroduction history and contemporary processes. Conserv. Genet. 11, 527–538 (2010).
Grossen, C., Biebach, I., Angelone-Alasaad, S., Keller, L. F. & Croll, D. Population genomics analyses of European ibex species show lower diversity and higher inbreeding in reintroduced populations. Evolut. Appl. 11, 123–139 (2018).
Brambilla, A., Biebach, I., Bassano, B., Bogliani, G. & von Hardenberg, A. Direct and indirect causal effects of heterozygosity on fitness-related traits in Alpine ibex. Proc. Biol. Sci. 282, 20141873 (2015).
Capparelli, R. et al. Protective effect of the Nramp1 BB genotype against Brucella abortus in the water buffalo (Bubalus bubalis). Infect. Immun. 75, 988–996 (2007).
Godfroid, J., Garin-Bastuji, B., Saegerman, C. & Blasco, J. M. Brucellosis in terrestrial wildlife. Revue Scientifique et Technique. Office International des Epizooties (2013).
Yang, A., Gomez, J. P., Haase, C. G., Proffitt, K. M. & Blackburn, J. K. Effects Of brucellosis serologic status on physiology and behavior of rocky mountain elk (Cervus canadensis nelsoni) in southwestern Montana. Journal of wildlife diseases (2018).
Schaschl, H., Wandeler, P., Suchentrunk, F., Obexer-Ruff, G. & Goodman, S. J. Selection and recombination drive the evolution of MHC class II DRB diversity in ungulates. Heredity 97, 427–437 (2006).
Grossen, C. et al. Introgression from domestic goat generated variation at the major histocompatibility complex of alpine ibex. PLoS Genet. 10, e1004438 (2014).
Zachos, F. E. et al. Genetic structure and effective population sizes in European red deer (Cervus elaphus) at a continental scale: insights from microsatellite DNA. J. Heredity 107, 318–326 (2016).
Biebach, I. & Keller, L. F. A strong genetic footprint of the re-introduction history of Alpine ibex (Capra ibex ibex). Mol. Ecol. 18, 5046–5058 (2009).
Keller, L. F. & Waller, D. M. Inbreeding effects in wild populations. Trends Ecol. Evolution 17, 230–241 (2002).
Saccheri, I. et al. Inbreeding and extinction in a butterfly metapopulation. Nat. 392, 491–494 (1998).
Brambilla, A., Keller, L., Bassano, B. & Grossen, C. Heterozygosity–fitness correlation at the major histocompatibility complex despite low variation in Alpine ibex (Capra ibex). Evolut. Appl. 11, 631–644 (2018).
Bateson, Z. W. et al. Specific alleles at immune genes, rather than genome-wide heterozygosity, are related to immunity and survival in the critically endangered Attwater’s prairie-chicken. Mol. Ecol. 25, 4730–4744 (2016).
Grueber, C. E. et al. Reciprocal translocation of small numbers of inbred individuals rescues immunogenetic diversity. Mol. Ecol. 26, 2660–2673 (2017).
Angelone, S. et al. Hidden MHC genetic diversity in the Iberian ibex (Capra pyrenaica). BMC Genet. 19, 28 (2018).
Hughes, A. L. & Yeager, M. Natural selection and the evolutionary history of major histocompatibility complex loci. Front. Biosci. 3, d509–516 (1998).
Werling, D., Jann, O., Offord, V., Glass, E. & Coffey, T. Variation matters: TLR structure and species-specific pathogen recognition. Trends Immunol. 30, 124–130 (2009).
Love, W., Dobbs, N., Tabor, L. & Simecka, J. W. Toll-like receptor 2 (TLR2) plays a major role in innate resistance in the lung against murine Mycoplasma. PLoS one 5, e10739 (2010).
Zuo, L., Wu, Y. & You, X. Mycoplasma lipoproteins and Toll-like receptors. J. Zhejiang Univ. Sci. B 10, 67 (2009).
Campos, M. A. et al. Role of Toll-like receptor 4 in induction of cell-mediated immunity and resistance to Brucella abortus infection in mice. Infect. Immun. 72, 176–186 (2004).
Oliveira, S. C., de Almeida, L. A., Carvalho, N. B., Oliveira, F. S. & Lacerda, T. L. Update on the role of innate immune receptors during Brucella abortus infection. Veterinary immunology immunopathology 148, 129–135 (2012).
Alim, M. A., Fu, Y., Wu, Z., Zhao, S. & Cao, J. Single Nucleotide Polymorphisms of Toll-Like Receptors and Association with Haemonchus contortus Infection in Goats. Pak. Veterinary J. 36, 286–291 (2016).
Mucha, R., Bhide, M., Chakurkar, E., Novak, M. & Mikula, I. Sr Toll-like receptors TLR1, TLR2 and TLR4 gene mutations and natural resistance to Mycobacterium avium subsp. paratuberculosis infection in cattle. Veterinary immunology immunopathology 128, 381–388 (2009).
Kloch, A. et al. Signatures of balancing selection in toll-like receptor (TLRs) genes–novel insights from a free-living rodent. Sci. Rep. 8, 8361 (2018).
Quemere, E. et al. Fluctuating pathogen-mediated selection drives the maintenance of innate immune gene polymorphism in a widespread wild ungulate. bioRxiv 458216 (2018).
Spurgin, L. & Richardson, D. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc. Biol. Sci. 277, 979–988 (2010).
Borriello, G. et al. Genetic resistance to Brucella abortus in the water buffalo (Bubalus bubalis). Infect. Immun. 74, 2115–2120 (2006).
Frankham, R. Challenges and opportunities of genetic approaches to biological conservation. Biol. Conserv. 143, 1919–1927 (2010).
Blanchong, J. A., Robinson, S. J., Samuel, M. D. & Foster, J. T. Application of genetics and genomics to wildlife epidemiology. J. Wildl. Manag. 80, 593–608 (2016).
McKnight, D. T., Schwarzkopf, L., Alford, R. A., Bower, D. S. & Zenger, K. R. Effects of emerging infectious diseases on host population genetics: a review. Conserv. Genet. 18, 1235–1245 (2017).
Coltman, D. W., Wilson, K., Pilkington, J. G., Stear, M. J. & Pemberton, J. M. A microsatellite polymorphism in the gamma interferon gene is associated with resistance to gastrointestinal nematodes in a naturally-parasitized population of Soay sheep. Parasitology 122, 571–582 (2001).
Maudet, C. et al. Microsatellite DNA and recent statistical methods in wildlife conservation management: applications in Alpine ibex [Capra ibex (ibex)]. Mol. Ecol. 11, 421–436 (2002).
Gauthier, D. & Villaret, J.-C. La réintroduction en France du bouquetin des Alpes. In Colloque sur les” Réintroductions Et Renforcements D’espèces Animales En France”, 6-8 décembre 1988, Saint-Jean-du-Gard, France, FRA (Société nationale de protection de la nature et d’acclimatation de France …, 1990).
Lambert, S. et al. High shedding potential and significant individual heterogeneity in naturally-infected Alpine ibex (Capra ibex) with Brucella melitensis. Front. Microbiology 9, 1065 (2018).
Hars, J. et al. Un foyer de brucellose chez les ongulés sauvages du massif du Bargy en Haute-Savoie. Bull. Epidemiol. Santé Anim. Alim 60, 2–6 (2013).
Van Oosterhout, C., Hutchinson, W. F., Wills, D. P. M. & Shipley, P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4, 535–538 (2004).
Vacca, G. M. et al. Chromosomal localisation and genetic variation of the SLC11A1 gene in goats (Capra hircus). Veterinary J. 190, 60–65 (2011).
Blackwell, J. M. et al. SLC11A1 (formerly NRAMP1) and disease resistance: Microreview. Cell. microbiology 3, 773–784 (2001).
Piry, S., Guivier, E., Realini, A. & Martin, J. SESAME Barcode: NGS-oriented software for amplicon characterization–application to species and environmental barcoding. Mol. Ecol. Resour. 12, 1151–1157 (2012).
Librado, P. & Rozas, J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinforma. 25, 1451–1452 (2009).
Belkhir, K., Borsa, P., Chikhi, L., Raufaste, N. & Bonhomme, F. GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. (1996).
Goudet, J. FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). (2001).
Coltman, D. W., Pilkington, J. G., Smith, J. A. & Pemberton, J. M. Parasite-Mediated Selection Against Inbred Soay Sheep In A Free-Living Island Populaton. Evolution 53, 1259–1267 (1999).
Stoffel, M. A. et al. inbreedR: an R package for the analysis of inbreeding based on genetic markers. Methods Ecol. Evolution 7, 1331–1339 (2016).
Anderson, D. R. Model based inference in the life sciences: a primer on evidence. (Springer Science & Business Media, 2007).
Burnham, K. & Anderson, D. Model Selection and Multi-Model Inference: a practical information- theoretic approach. (Springer-Verlag, 2002).
Symonds, M. R. & Moussalli, A. A brief guide to model selection, multimodel inference and model averaging in behavioural ecology using Akaike’s information criterion. Behav. Ecol. Sociobiol. 65, 13–21 (2011).
Zuur, A., Ieno, E. N., Walker, N., Saveliev, A. A. & Smith, G. M. Mixed effects models and extensions in ecology with R. (Springer Science & Business Media, 2009).
Bates, D., Maechler, M. & Bolker, B. lme4: Linear mixed-effects models using S4 classes. (2012).
Barton, K. MuMIn: multi-model inference. R package version 1. 0. 0. http://r-forge.r-project. org/projects/mumin/ (2009).
Mazerolle, M. J. AICcmodavg: Model selection and multimodel inference based on (Q) AIC (c). R. package version 1, 35 (2013).
Acknowledgements
The authors would like to thank Carole Toïgo, the field technicians and researchers from the French Hunting and Wildlife Agency (Office National de la Chasse et de la Faune Sauvage, now Office Français de la Biodiversité) who performed ibex captures, monitoring and sampling. We also thank the local veterinary services, the local veterinarians and the Préfecture of Haute Savoie who supported the field work and the departmental laboratory staff that performed blood DNA extractions. Data used in this work were partly produced through the genotyping and sequencing facilities of ISEM (Institut des Sciences de l’Evolution-Montpellier) and LabEx Centre Méditerranéen Environnement et Biodiversité. Thanks also go to Lukas Keller and Alice Brambilla for helpful suggestions and comments. We lastly thank the Ministry of Agriculture and Food, the French Agency for Food, Environmental and Occupational Health & Safety (ANSES) and the French Hunting and Wildlife Agency who funded this study.
Author information
Authors and Affiliations
Contributions
E.Q., S.R., E.G.F. designed the study and interpreted the data; E.P., P.M. and S.R. collected the data on the field. M.G., Y.G. and J.M. handled the laboratory work; E.Q. performed the population genetic and statistical analyses; E.Q. and E.G.F. led the writing, all authors contributed to the manuscript and gave final approval for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Quéméré, E., Rossi, S., Petit, E. et al. Genetic epidemiology of the Alpine ibex reservoir of persistent and virulent brucellosis outbreak. Sci Rep 10, 4400 (2020). https://doi.org/10.1038/s41598-020-61299-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-61299-2
This article is cited by
-
Deletion in KARLN intron 5 and predictive relationship with bovine tuberculosis and brucellosis infection phenotype
Veterinary Research Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.