Abstract
Two new cyclopentapeptides, xylapeptide A (1) with an uncommon L-pipecolinic acid moiety, and xylapeptide B (2) having a common L-proline residue were identified from an associated fungus Xylaria sp. isolated from the Chinese medicinal plant Sophora tonkinensis. Their planar structures were elucidated by a comprehensive analysis of NMR and MS spectroscopic spectra. The absolute configurations were determined by Marfey’s method and single-crystal X-ray diffraction (Cu Kα) analysis. Xylapeptide A (1) is the first example of cyclopentapeptide with L-Pip of terrestrial origin and showed strong antibacterial activity against Bacillus subtilis and B. cereus with MIC value of 12.5 μg/mL.
Similar content being viewed by others
Introduction
New antibiotics are in need of development against the resistant microorganisms in controlling bacterial infections1, 2. Endophytes are increasingly attracting attention as sources of potentially valuable compounds3. The secondary metabolites produced by endophytes have received enduring development for chemical diversity and antimicrobial activities2, 4. Peptides are investigated intensively because they are target specific with higher degree of interactions5. In recent years, a quite number of peptides isolated from endophytes have been reported. It is noteworthy that peptides with good antimicrobial activities were isolated mainly from Streptomyces genus, such as kakadumycin A6 showed potent antibacterial activity against Staphylococcus aureus, S. simulans, S. pneumonia, and Shigella dysenteriae; munumbicin C7 displayed strong activity against vancomycin-sensitive S. aureus; actinomycin D8 exhibited strong activity against methicillin-resistant S. aureus and multidrug-resistant Mycobacterium tuberculosis, and munumbicin E-48 demonstrated strong activity against S. aureus. However, few peptides isolated from fungi exhibited good antimicrobial activities, the example was the cyclo-(Pro-Thr) and cyclo-(Pro-Tyr)9 (produced by a Penicillium sp. isolated the mangrove Acrostichum aureurm) showed a strong antibacterial activity against S. aureus and antifungal activity against Candida albicans.
Sophora tonkinensis is an important traditional Chinese medicinal plant used widely to treat acute pharyngolaryngeal infections and sore throats10, 11. In the course of our studies to investigate antimicrobial constituents from associated fungi, the fungus Xylaria sp. (GDG-102) was isolated from S. tonkinensis. The crude extract of the fermentation broth exhibited antibacterial activity. Preliminary UPLC-MS and 1H NMR analysis showed that this fungus produced relatively polar peptide compounds (Figure S1). Chemical investigation led to the identification of two new cyclopentapeptides, xylapeptides A (1) and B (2) (Fig. 1). Xylapeptide A is the first cyclopentapeptide with L-Pip of terrestrial origin, and exhibited strong antibacterial activity against Bacillus subtilis and B. cereus. Xylapeptide B showed strong antibacterial activity against B. subtilis, B. cereus, B. megaterium, Micrococcus luteus, S. aureus, Shigella castellani, as well as strong antifungal activity against C. albicans. Herein, we report the structure determination and biological activities of the two cyclic peptides.
Results and Discussion
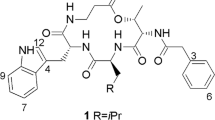
Xylapeptide A (1) was obtained as a colorless crystal. Its HRESIMS gave a [M + H]+ at m/z 556.3505, indicating a molecular formula of C30H45N5O5 and requiring 11 degrees of unsaturation. The 1H NMR spectrum (Table 1) suggested that it has three amide (NH) protons at δ H 7.45, 7.11, and 6.13, one N-methyl group at δ H 2.97, and four α-amino protons at δ H 4.74, 4.61, 4.34, and 4.14. It also showed a single mono-substituted benzene at δ H 7.31, 7.26 and 7.17. The 13C NMR spectrum indicated that it has five amide type carbonyls at δ C 172.8, 172.6, 171.8 (2 C) and 170.2, six nitrogenated sp3 carbon resonance at δ C 64.2, 59.0, 57.0, 48.1, 47.5 and 47.4. These above NMR features accounted for 9 of the 11 unsaturations. Further analysis of 2D NMR spectrum allowed five subunits to be established, an N-Me-phenylalanine (N-Me-Phe), a valine (Val), an alanine (Ala), a leucine (Leu), and a pipecolinic acid (Pip) (Fig. 2). Compound 1 was concluded to be a cyclopeptide on the basis of these spectral characteristics.
The sequence of residues in 1 was elucidated on the basis of HMBC correlations (Fig. 2). The HMBC correlations from Pip-H-6 to Leu-CO, Leu-NH to Ala-CO, Ala-NH to Val-CO, Val-NH to N-Me-Phe-CO, and N-Me-Phe-α-H to Pip-CO allowed the definition of the residue sequence of 1 as cyclo-(CO-Pip → Leu → Ala → Val → N-Me-Phe-N). Furthermore, the ESI MS2 (Fig. S16) fragment ions corresponding to neutral losses of [Pip], [Pip−Leu], [Pip−Leu−Ala], and [Pip−Leu−Ala−Val] were also observed which confirmed the cyclic structure for 1. The configurations of the amino acid residues of 1 was determined by acid hydrolysis, derivatization with N α-(2,4-dinitro-5-fluorophenyl)-L-alalinamide (L-FDAA, the advanced Marfey’s method12), and UPLC-MS analysis of the derivatives with comparison to the standards (Figure S17). Retention times (min) of the standard amino acid derivatives were as follows: L-Ala, D-Ala, L-Val D-Val, L-Leu, D-Leu, N-Me-L-Phe and N-Me-D-Phe were 5.817, 6.811, 7.692, 8.977, 8.996, 10.212, 9.281, and 9.736 min, respectively. Retention times of the derivatives of the acid hydrolysate of 1 were 6.793, 7.676, 8.975, and 9.263 min. It indicated that Leu, Val, and N-Me-Phe had the L-configuration, while Ala was D-configured. Finally, the absolute configuration of 1 was confirmed unambiguously by single-crystal X-ray analysis which indicated an L-configuration of Pip (Fig. 3).
Xylapeptide B (2) was also obtained as a colorless crystal. Its molecular formula C29H43N5O5 (eleven degrees of unsaturation) was determined on the basis of HRESIMS data. The 1H and 13C NMR spectrum (Table 2) indicated that 2 was very similar to 1 with the additional high-field resonances (δ H 0.79/1.57, δ C 23.2 (CH2)) in 1 were absent. Examination of the 2D NMR spectra revealed that 2 shared the same N-Me-Phe, Val, Ala, and Leu units with 1, the only difference is that the Pip in 1 was replaced by a proline (Pro) subunit in 2. The sequence of the amino acids of 2 was also assigned by HMBC correlations (Fig. 4) and fully supported by the ESI MS2 experiment results (Fig. S16) which revealed the same linkage with 1. The configurations of the amino acid residues were also determined by Marfey’s method. The retention times (min) of the standard L-Pro and D-Pro were 6.262 and 6.689 min, respectively. The retention times for the acid hydrolysate of 2 were 6.285, 6.818, 7.707, 9.004, and 9.318 min, respectively, which correspond to L-Pro, L-Leu, L-Val, N-Me-L-Phe, and D-Ala (Figure S17). Besides, by slow crystallization from CH3OH, single crystals of 2 suitable for X-ray diffraction analysis using Cu Kα radiation was also obtained, allowing the complete structure to be established unambiguously (Fig. 3).
Naturally occurring cyclic peptides with nonproteinogenic amino acids mainly contained pipecolinic acid (Pip) and anthranilic acid (Ant)13. Up to now, nearly twenty cyclic peptides with Pip residues isolated from nature sources have been reported which occupied a very small proportion of the identified peptide. Most of the cyclic peptides with Pip residues reported so far were of a marine origin14, 15 such as cyclopentadepsipeptides JBIR-113, 114 and 115 (fungus Penicillium sp16.), apratoxin H (cyanobacterium Moorea producens 17), petrosifungins A and B (fungus Penicillium brevicompactum 18), cyclohexapeptide similanamide (fungus Aspergillus similanensis 19), cyclohexadepsipeptides somamides A and B (cyanobacteria Lyngbya majuscule and Schizothrix sp20.), symplostatins (cyanobacterium Symploca hydnoides 21), tasipeptins A and B (cyanobacterium Symploca sp22.), kurahamide (cyanobacterium Moorea sp23.), and dolastatins (sea hare Dolabella auricularia 24), cycloheptadepsipeptides neamphamides B−D (sponge Neamphius huxleyi 25), cyclooctadepsipeptide homophymine A (sponge Homophymia sp26.), and pipecolidepsins A and B (sponge Homophymia lamellose 27). Cyclic peptides with Pip from terrestrial origins are rare including cyclohexapeptides PF1171 A−G (unknown ascomycete OK-12813), and cyclodecadepsipeptides, clavariopsins A and B (aquatic hyphomycetes Clavariopsis aquatic 28). Up to now, the Pip in all the peptides is L-configuration. Xylapeptide A (1) is the first cyclopentapeptide from a terrestrial origin with L-Pip.
The transformation of Lys to Pip in plants and fungus has been studied by radioactive labeling experiment. In the Ca-deficient wheat plants, the 14C-L-Lys was transported rapidly in to the shoots and then degraded to Pip in many pathways29, 30. In the Rhizoctonia leguminicola fungus, the L-[U-14C]-Lys is used to form L-Pip31, 32. Although Gatto and co-workers reported the first in vitro characterization of a lysine cyclodeaminase (RapL) which utilized the NAD+ as a cofactor in a catalytic manner in Streptomyces hygroscopicus to catalyst lysine into Pip33, the genes controlling the biosynthesis of Lys to Pip have not been described yet. It is suggested that the Pip in xylapeptide A (1) may also be converted from lysine, and a radioactive labeling experiment is needed.
Xylapeptides A (1) and B (2) were evaluated for their antibacterial activity. Compound 1 showed strong and selective antibacterial activity against Bacillus subtilis and B. cereus with MIC value of 12.5 μg/mL. Compound 2 exhibited strong and broad antibacterial spectrum against B. subtilis, B. cereus, B. megaterium, Micrococcus luteus, Staphylococcus aureus, and Shigella castellani with MIC values of 12.5, 6.25, 6.25, 12.5, 12.5, and 12.5 μg/mL, respectively. Compound 2 also showed a strong antifungal activity against Canidia albicans with MIC value of 12.5 μg/mL. This suggested that the L-Pip residue and L-Pro residue play different roles in the antimicrobial activity. The cytotoxic, α-glucosidase inhibitory, and antiviral activities of 1 and 2 were also evaluated. However, they were all inactive.
In summary, two new cyclopentapeptides, xylapeptides A (1) and B (2), were isolated from a Xylaria sp. fungus cultured from the Chinese medicinal plant S. tonkinensis. Xylapeptide A (1) contains a non-proteinogenic amino acid, L-pipecolinic acid (L-Pip), which is the first example of cyclopentapeptide from a terrestrial origin with L-Pip. Compound 1 displayed a strong antibacterial activity against Bacillus subtilis and B. cereus. Xylapeptide B (2) has a broader antibacterial spectrum as well as antifungal activity.
Methods
General experimental procedures
Optical rotations were measured on a JASCO P-1020 digital polarimeter. UV spectra were determined on a Shimadzu double-beam 210 A spectrometer. IR spectra were taken on a Bruker EQUINOX 55 spectrometer using KBr pellets. 1D and 2D NMR spectra were obtained on an Agilent DD2 400 MHz NMR spectrometer or a DRX-500 spectrometer, respectively, with TMS as the internal standard. ESIMS and HRESIMS spectra were obtained from a Micromass Q-TOF spectrometer and a Thermo Scientific LTQ Orbitrap XL spectrometer. Semi-preparative HPLC was performed on a Waters 1525 system using a C18 (Kromasil, 5 μm, 10 × 250 mm) column coupled with a Waters 2996 photodiode array detector. UPLC MS was performed on Waters UPLC® system using a C18 column [ACQUITY UPLC® BEH C18, 2.1 × 50 mm, 1.7 μm; 0.5 mL/min] and ACQUITY QDa ESIMS scan from 150 to 1000 Da. Silica gel (Qing Dao Hai Yang Chemical Group Co.; 200–300 mesh), and octadecylsilyl silica gel (Unicorn; 45–60 μm), were used for column chromatography (CC). Precoated silica gel plates (Yan Tai Zi Fu Chemical Group Co.; G60, F-254) were used for thin-layer chromatography.
Isolation and identification of the fungal material
The fungus Xylaria sp. (strain # GDG-102) was isolated from the Chinese medicinal plant S. tonkinensis collected in Hechi, Guangxi Province, in October, 2014. The fungus Xylaria sp. (strain # GDG-102) was isolated from fresh healthy leaves of Chinese medicinal plant S. tonkinensis collected in Hechi, Guangxi Province, in October, 2014. The leaves were designed to undergo a process described as surface sterilization34. The surface-sterilized leaves were aseptically sectioned into small pieces and plated onto the PDA plates containing an antibiotic to suppress bacterial growth (composition of isolation medium: potatoes 200 g/L, glucose 20 g/L, agar 15 g/L and ampicillin sodium 0.2 g/L in water). After incubation at 27 °C, the fungal strain under investigation was isolated from the growing cultures by repeated reinoculation on PDA plates. The fungus was identified as Xylaria sp. (GenBank accession number KU645984) using a molecular biological protocol by DNA amplification and sequencing of the ITS region as described in ref. 34.
Fermentation, extraction, and isolation
The fungal strain was cultivated in 20 L of liquid medium (composition of medium: 200 g/L cooked and sliced potatoes, 20 g/L glucose in water, in 1 L Erlenmeyer flasks each containing 500 mL of culture broth) at room temperature without shaking for 7 weeks. Then the culture was filtered to separate the culture broth from the mycelia. The culture broth was extracted with an equal volume of ethyl acetate (EtOAc) and the fungal mycelia were extracted with CH2Cl2-CH3OH (v:v, 1:1) for three times, respectively. The mycelia extraction were concentrated to about 0.3 L and extracted with EtOAc. The EtOAc layer was evaporated to dryness under reduced pressure to give broth extract (4.5 g) and mycelia extract (4.8 g), respectively. The broth extract was subjected to silica gel CC (petroleum ether (PE)−EtOAc, v-v, gradient) to afford five fractions (Fr.1–Fr.5). Fr.4 was repeated purified by ODS CC (CH3OH−H2O) and then recrystallized in MeOH to obtain 2 (20 mg). In search for similar compounds, the mycelia extraction was subjected to silica gel CC (PE−EtOAc, v:v, gradient) to afford six fractions (Fr.1–Fr.6). Under the guidance of 1H NMR, Fr.4 was chromatographed on ODS with CH3OH−H2O, and then purified by semi-preparative HPLC (CH3OH-H2O, v:v, 83:17) to yield 1 (10 mg).
Xylapeptide A ( 1 ). colorless crystals; \({[\alpha ]}_{{\rm{D}}}^{20}\) −15.6 (c 0.1, CH3OH); UV (CH3OH) λ max (log ε) 205 (4.44) nm; IR (KBr) ν max 3446, 1683, 1521 cm−1; 1H and 13C NMR data, see Table 1. ESI MS2 (fragmentation of m/z 556.4 [M + H]+) m/z 445.2 [M − Pip + H]+, 332.2 [M − Pip − Leu + H]+, 261.1 [M − Pip − Leu − Ala + H]+, 162.1 [M − Pip − Leu − Ala − Val + H]+; HRESIMS m/z 556.3505 [M + H]+ (calcd for C30H46N5O5, 556.3493).
Xylapeptide B ( 2 ). colorless crystals; \({[\alpha ]}_{{\rm{D}}}^{20}\) − 85.7 (c 0.2, CH3OH); UV (CH3OH) λ max (log ε) 206 (4.47) nm; IR (KBr) ν max 3434, 2959, 1648, 1523, 1454 cm−1; 1H and 13C NMR data, see Table 2; ESI MS2 (fragmentation of m/z 542.3 [M + H]+) m/z 443.2 [M − Val + H]+, 372.2 [M − Val − Ala + H]+, 259.1 [M − Val − Ala − Leu + H]+, 162.0 [M − Val − Ala − Leu − Pro + H]+; HRESIMS m/z 542.3345 [M + H]+ (calcd for C29H44N5O5, 542.3337).
X-ray crystallographic analysis of compounds 1 and 2
Upon crystallization from CH3OH-H2O, the regular needle crystals of 1 and 2 were obtained. The single-crystal X-ray diffraction data were collected at 293(2) K for 1 and 296.15 K for 2 on a Bruker APEX-II CCD diffractometer with Cu Kα radiation (λ = 1.54178 Å). Crystallographic data for 1 and 2 have been deposited with the Cambridge Crystallographic Data Centre. Copies of the data can be obtained, free of charge, on application to the Director, CCDC, 12 Union Road, Cambridge CB2 1EZ, UK (fax: + 44-(0)1223-336033, or e-mail: deposit@ccdc.cam.ac.uk).
Crystal data for 1: C30H45N5O5∙2(H2O) Mr = 591.74, monoclinic, space group P2(1) with a = 13.3173 (2) Å, b = 6.90755 (8) Å, c = 17.8276 (3) Å, α = 90.00°, β = 93.7299 (14)°, γ = 90.00°, V = 1636.50 (4) Å3, Z = 2, D x = 1.201 mg/m3, μ (Cu Kα) = 1.54178 mm−1, and F(000) = 640. Crystal dimensions: 0.40 × 0.28 × 0.10 mm3. Independent reflections: 20526, the final R 1 value was 0.0337, wR 2 = 0.0824 (I > 2σ(I)), Flack parameter = 0.04(15). CCDC number: 1501650.
Crystal data for 2: C29H43N5O5∙H2O∙CH3OH, Mr = 591.74, monoclinic, space group P2(1) with a = 9.9670 (5) Å, b = 12.5923(7) Å, c = 26.2574 (14) Å, α = 90.00°, β = 90.00°, γ = 90.00°, V = 3295.5(3) Å3, Z = 4, D x = 1.193 mg/m3, μ (Cu Kα) = 1.54184 mm−1, and F(000) = 1280. Crystal dimensions: 0.30 × 0.15 × 0.08 mm3. Independent reflections: 20526, The final R 1 value was 0.0756, wR 2 = 0.1010 (I > 2σ(I)), Flack parameter = −0.1(2). CCDC number: 1502010.
Absolute configuration of xylapeptides A (1) and B (2)
The amino acid standards (L- and D/L-configurations) relevant to 1 and 2 were obtained commercially and 0.5 mg was dissolved in 20 μL H2O. Each standard was then derivatized for Marfey’s analysis by adding 1 M NaHCO3 (10 μL) and L-FDAA (1% w/v in acetone, 50 μL). The mixture was heated at 45 °C for 1 h with continuous stirring, then neutralized with 1 M HCl after cooling at room temperature. The derivatives were then dried and diluted with CH3OH and analyzed by UPLC MS (using a C18 column [ACQUITY UPLC® BEH C18, 2.1 × 50 mm, 1.7 μm; 0.5 mL/min, UV detection at 340 nm; ACQUITY QDa ESIMS scan from 150 to 1000 Da; linear gradient: (A) CH3CN and (B) H2O with 0.1% HCOOH 5–50% A (0–13 min), 50–100% A (13–15 min), 100% A (15–17 min), 100–5% A (17–18 min), 5% A (18–20 min). Retention times (min) of the standard amino acid derivatives of L-Ala, D-Ala, L-Pro, D-Pro, L-Val D-Val, L-Leu, D-Leu, N-Me-L-Phe and N-Me-D-Phe were 5.817, 6.811, 6.262, 6.689, 7.692, 8.977, 8.996, 10.212, 9.281, and 9.736 min, respectively.
Xylapeptide A (or xylapeptide B, 0.5 mg) was hydrolyzed with 6 M HCl (2.0 mL) at 110 °C for 15 h. The solution was evaporated to dryness, and derivatized for Marfey’s analysis in a similar manner to the derivatized standard amino acids. The derivatives of the acid hydrolysate of 1 were analyzed by UPLC-MS with the retention times as follows: 6.793 (D-Ala), 7.676 (L-Val), 8.975 (L-Leu), and 9.263 (N-Me-L-Phe) min. The retention times of that in 2 were: 6.285 (L-Pro), 6.818 (D-Ala), 7.707 (L-Val), 9.004 (L-Leu), and 9.318 (L-N-Me-Phe) min.
Biological assays
The antibacterial activity against B. subtilis, B. cereus, B. megaterium, Micrococcus luteus, Staphylococcus aureus, and Shigella castellani, and antifungal activity against C. albicans were evaluated by using 96-well microtitre plates35, with ampicillin as positive control. The cytotoxic activity against human colon carcinoma (HCT-116), human cervical cancer (HeLa), non-small cell lung carcinoma (A549), human breast cancer (MCF-7), human pancreatic carcinoma (BXPC-3), and chronic myelocytic leukemia (K562) cell lines was evaluated by the SRB method36 and MTT method37, with adriamycin as a positive control. The antiviral activity against human cytomegalovirus (HCMV), and herpes simplex virus type 1 (HSV-1) virus was evaluated by the cytopathic effect inhibition assay38, with cidofovir and acyclovir as positive control. The α-glucosidase inhibitory activity was evaluated by Yilmazer-Musa’s method39, with acarbose as a positive control.
References
Alvin, A., Miller, K. I. & Neilan, B. A. Exploring the Potential of Endophytes from Medicinal Plants as Sources of Antimycobacterial Compounds. Microbiol. Res. 169, 483–495 (2014).
Golinska, P. et al. Endophytic Antinobacteria of Medicinal Plants: Diversity and Bioactivity. Anton. Van. Leeuwenhoek 108, 267–289 (2015).
Martinez-Klimova, E., Rodríguez-Peña, K. & Sánchez, S. Endophytes as Sources of Antibiotics. Biochem. Pharmacol. doi:10.1016/j.bcp.2016.10.010 (2016).
Zhang, H. W., Song, Y. C. & Tan, R. X. Biology and Chemistry of Endophytes. Nat. Prod. Rep. 23, 753–771 (2006).
Abdalla, M. A. & Matasyoh, J. C. Endophytes as Producers of Peptides: An overview about the Recently Discovered Peptides from Endophytic Microbes. Nat. Prod. Bioprospect. 4, 257–270 (2014).
Castillo, U. et al. Kakadumycins, Novel Antibiotics from Streptomyces sp. NRRL 30566, an Endophyte of Grevillea pteridifolia. FEMS Microbiol. Lett. 224, 183–190 (2003).
Castillo, U. et al. Munumbicins, Wide-Spectrum Antibiotics Produced by Streptomyces NRRL 30562, Endophytic on Kennedia nigriscans. Microbiology 148, 2675–2685 (2002).
Castillo, U. et al. Munumbicins E-4 and E-5: Novel Broad-Spectrum Antibiotics from Streptomyces NRRL 3052. FEMS Microbiol. Lett. 255, 296–300 (2006).
Cui, H. B. et al. Antibacterial Constituents from the Endophytic Fungus Penicillium sp. 0935030 of Mangrove Plant Acrostichum aureum. Zhongguo Kangshengsu Zazhi 33, 407–410 (2008).
China Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China. Chemical Industry Press, Beijing (2005).
Zhang, Y. S. et al. Research Progress of Biological Activity of Vietnamese Sophora Root. Zhongyiyao Xuebao 41, 96–97 (2013).
Marfey, P. Determination of D-amino acids. II. Use of a Bifunctional Reagent, 1,5-Difluoro-2,4-dinitrobenzene. Carlsberg Res. Commun. 49, 591–596 (1984).
Masuda, Y. et al. Structure Revision of Similanamide to PF1171C by Total Synthesis. J. Nat. Prod. 78, 2286–2291 (2015).
Pan, C., Al-Kareef, A. M. Q. & Wang, H. Cyclopeptides from Marine Organisms. Mini-Reviews Org. Chem. 12, 196–214 (2015).
Blunt, J. W. et al. Marine Natural Products. Nat. Prod. Rep. 33, 382–431 (2016).
Kawahara, T., Takagi, M. & Shin-ya, K. Three New Depsipeptides, JBIR-113, JBIR-114 and JBIR-115, Isolated from a Marine Sponge-Derived Penicillium sp. fS36. J. Antibiot. 65, 147–150 (2012).
Thornburg, C. C. et al. Apratoxin H and Apratoxin A Sulfoxide from the Red Sea Cyanobacterium Moorea producens. J. Nat. Prod. 76, 1781–1788 (2013).
Bringmann, G. et al. Petrosifungins A and B, Novel Cyclodepsipeptides from a Sponge-Derived Strain of Penicillium brevicompactum. J. Nat. Prod. 67, 311–315 (2004).
Prompanya, C. et al. A New Cyclic Hexapeptide and a New Isocoumarin Derivative from the Marine Sponge-Associated Fungus Aspergillus similanensis KUFA 0013. Mar. Drugs 13, 1432–1450 (2015).
Nogle, L. M. et al. Somamides A and B, two New Depsipeptide Analogues of Dolastatin 13 from a Fijian Cyanobacterial Assemblage of Lyngbya majuscule and Schizothrix species. J. Nat. Prod. 64, 716–719 (2001).
Harrigan, G. G. et al. Symplostatin 2: a Dolastatin 13 Analogue from the Marine Cyanobacterium Symploca hydnoides. J. Nat. Prod. 62, 655–658 (1999).
Williams, P. G. et al. Tasipeptins A and B: New Cytotoxic Depsipeptides from the Marine Cyanobacterium Symploca sp. J. Nat. Prod. 66, 620–624 (2003).
Iwasaki, A. et al. Kurahamide, a Cyclic Depsipeptide Analog of Dolastatin 13 from a Marine Cyanobacterial Assemblage of Lyngbya sp. Bull. Chem. Soc. Japan. 87, 609–613 (2014).
Prttit, G. R. et al. Isolation and Structure of the Cytostatic Depsipeptide Dolastatin 13 from the Sea Hare Dolabella auricularia. J. Am. Chem. Soc. 11, 5015–5017 (1989).
Coello, L. Isolation and Structures of Pipecolidepsins A and B, Cytotoxic Cyclic Depsipeptides from the Madagascan Sponge Homophymia lamellose. J. Nat. Prod. 77, 298–303 (2014).
Tran, T. D. et al. Cytotoxic Cyclic Depsipeptides from the Australian Marine Sponge Neamphius huxleyi. J. Nat. Prod. 75, 2200–2208 (2012).
Zampella, A. et al. Homophymine A, an Anti-HIV Cyclodepsipeptide from the Sponge Homophymia sp. J. Org. Chem. 73, 5319–5327 (2008).
Suzuki, Y. et al. New Cyclic Depsipeptide Antibiotics, Clavariopsins A and B, Produced by an Aquatic Hyphomycetes, Clavariopsis aquatica. 2. Structure Analysis. J. Antiboit. 54, 22–28 (2011).
Rothstein, M. & Miller, L. L. The Conversion of Lysine to Pipecolic Acid in the Rat. J. Biol. Chem. 211, 851–858 (1954).
ČinčerovÁ, A. & ČernÁ, E. Biosynthesis of Pipecolic Acid in Calcium-Deficient Wheat Plants. Z. Pflanzenphysiol. Bd. 74, 366–370 (1974).
Guengerich, F. P. & Broquist, H. P. Biosynthesis of Slaframine, (1S,6S,8aS)-1-acetoxy-6-aminooctahydroindolizine, a Parasympathomimetic Alkaloids of Fungal Origin. II. Origin of Pipecolic Acid. Biochemistry 12, 4270–4274 (1973).
Wickwire, B. M. et al. Pipecolic Acid Biosynthesis in Rhizoctonia leguminicola. J. Biol. Chem. 265, 14742–14747 (1990).
Gatto, G. J. et al. Biosynthesis of Pipecolic Acid by RapL, a Lysine Cyclodeaminase Encoded in the Rapamycin Gene Cluster. J. Am. Chem. Soc. 128, 3838–3847 (2006).
Aly, A. H. et al. Cytotoxic Metabolites from the Fungal Endophyte Alternaria sp. and Their Subsequent Detection in Its Host Plant Polygonum senegalense. J. Nat. Prod. 71, 972–980 (2008).
Appendio, G. et al. Antibacterial Cannabinoids from Cannabis sativa: a Structure-Activity Study. J. Nat. Prod. 71, 1427–1430 (2008).
Skehan, P. et al. New Colorimetric Cytotoxicity Assay for Anticancer-Drug Screening. J. Natl. Cancer Inst. 82, 1107–1112 (1990).
Mosmann, T. Rapid Colorimetric Assay for Cellular Growth and Survival: Application to Proliferation and Cytotoxicity assays. Immunol. Methods 65, 55–63 (1983).
Grassauer, A. et al. Concurrent Infections by All Four Dengue Virus Serotypes During an Outbreak of Dengue in 2006 in Delhi, India. Virol. J. 5, 1–13 (2008).
Yilmazer-Musa, M. et al. Grape Seed and Tea Extracts and Catechin 3-Gallates Are Potent Inhibitors of α-Amylase and α-Glucosidase Activity. J. Agric. Food Chem. 60, 8924–8929 (2012).
Acknowledgements
We thank Prof. Yu-Cheng Gu (Syngenta) for his proofreading of the manuscript and Syngenta for the fellowship to X.M.H. This work was supported by the Program for Innovative Research Team (No. IRT1225), the Guangxi Natural Science Foundation of China (No. 2014GXNSFAA118055), the Project of State Key Laboratory for the Chemistry and Molecular Engineering of Medicinal Resources (Guangxi Normal University), Ministry of Education of China (Nos. CMEMR2016-A06 and CMEMR2016-B04), Aoshan Talents Program Supported by Qingdao National Laboratory for Marine Science and Technology (No. 2015ASTP-ES11) and the Taishan Scholars Program, China.
Author information
Authors and Affiliations
Contributions
W.F.X. and X.M.H. contributed to extraction, isolation, identification, and manuscript preparation. F.H.Y. and N.Z. contributed to bioactivities test. J.L. and C.Y.W. contributed to NMR analysis. C.L.S. contributed to X-ray analysis. R.Y.Y. and C.L.S. conceived of and proposed the idea.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xu, WF., Hou, XM., Yao, FH. et al. Xylapeptide A, an Antibacterial Cyclopentapeptide with an Uncommon L-Pipecolinic Acid Moiety from the Associated Fungus Xylaria sp. (GDG-102). Sci Rep 7, 6937 (2017). https://doi.org/10.1038/s41598-017-07331-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-07331-4
This article is cited by
-
Secondary Metabolites from the Endophytic Fungus Xylaria sp. GDG-102 by Osmac Strategy and Their Antibacterial Activities
Chemistry of Natural Compounds (2024)
-
Secondary Metabolites from the Endophytic Fungus Xylaria grammica and Their Anti-Inflammatory Activities
Chemistry of Natural Compounds (2023)
-
Secondary Metabolites from the Fungus Metarhizium marquandii
Chemistry of Natural Compounds (2023)
-
Recent progress in biodiversity research on the Xylariales and their secondary metabolism
The Journal of Antibiotics (2021)
-
Secondary Metabolites of the Endophytic Fungus Xylaria sp. GDG-102 from Sophora tonkinensis
Chemistry of Natural Compounds (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.