Abstract
SETD5 gene mutations have been identified as a frequent cause of idiopathic intellectual disability. Here we show that Setd5-haploinsufficient mice present developmental defects such as abnormal brain-to-body weight ratios and neural crest defect-associated phenotypes. Furthermore, Setd5-mutant mice show impairments in cognitive tasks, enhanced long-term potentiation, delayed ontogenetic profile of ultrasonic vocalization, and behavioral inflexibility. Behavioral issues are accompanied by abnormal expression of postsynaptic density proteins previously associated with cognition. Our data additionally indicate that Setd5 regulates RNA polymerase II dynamics and gene transcription via its interaction with the Hdac3 and Paf1 complexes, findings potentially explaining the gene expression defects observed in Setd5-haploinsufficient mice. Our results emphasize the decisive role of Setd5 in a biological pathway found to be disrupted in humans with intellectual disability and autism spectrum disorder.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Mefford, H. C., Batshaw, M. L. & Hoffman, E. P. Genomics, intellectual disability, and autism. N. Engl. J. Med. 366, 733–743 (2012).
Rauch, A. et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 380, 1674–1682 (2012).
Najmabadi, H. et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 478, 57–63 (2011).
Dillon, S. C., Zhang, X., Trievel, R. C. & Cheng, X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 6, 227 (2005).
Grozeva, D. et al. De novo loss-of-function mutations in SETD5, encoding a methyltransferase in a 3p25 microdeletion syndrome critical region, cause intellectual disability. Am. J. Hum. Genet. 94, 618–624 (2014).
Kuechler, A. et al. Loss-of-function variants of SETD5 cause intellectual disability and the core phenotype of microdeletion 3p25.3 syndrome. Eur. J. Hum. Genet. 23, 753–760 (2015).
Powis, Z. et al. Expansion and further delineation of the SETD5 phenotype leading to global developmental delay, variable dysmorphic features, and reduced penetrance. Clin. Genet. 93, 752–761 (2018).
Green, C., Willoughby, J. & Balasubramanian, M. De novo SETD5 loss-of-function variant as a cause for intellectual disability in a 10-year old boy with an aberrant blind ending bronchus. Am. J. Med. Genet. A. 173, 3165–3171 (2017).
Osipovich, A. B., Gangula, R., Vianna, P. G. & Magnuson, M. A. Setd5 is essential for mammalian development and the co-transcriptional regulation of histone acetylation. Development 143, 4595–4607 (2016).
Baxter, L. L., Hou, L., Loftus, S. K. & Pavan, W. J. Spotlight on spotted mice: a review of white spotting mouse mutants and associated human pigmentation disorders. Pigment Cell Res. 17, 215–224 (2004).
Cordero, D. R. et al. Cranial neural crest cells on the move: their roles in craniofacial development. Am. J. Med. Genet. A. 155A, 270–279 (2011).
Santos, A. R., Kanellopoulos, A. K. & Bagni, C. Learning and behavioral deficits associated with the absence of the Fragile X mental retardation protein: what a fly and mouse model can teach us. Learn. Mem. 21, 543–555 (2014).
Deacon, R. M. Assessing nest building in mice. Nat. Protoc. 1, 1117–1119 (2006).
Moretti, P., Bouwknecht, J. A., Teague, R., Paylor, R. & Zoghbi, H. Y. Abnormalities of social interactions and home-cage behavior in a mouse model of Rett syndrome. Hum. Mol. Genet. 14, 205–220 (2005).
Wöhr, M. Ultrasonic vocalizations in Shank mouse models for autism spectrum disorders: detailed spectrographic analyses and developmental profiles. Neurosci. Biobehav. Rev. 43, 199–212 (2014).
Thomas, A. et al. Marble burying reflects a repetitive and perseverative behavior more than novelty-induced anxiety. Psychopharmacology (Berl.) 204, 361–373 (2009).
Wolmarans, W., Stein, D. J. & Harvey, B. H. Of mice and marbles: novel perspectives on burying behavior as a screening test for psychiatric illness. Cogn. Affect. Behav. Neurosci. 16, 551–560 (2016).
Moy, S. S. et al. Development of a mouse test for repetitive, restricted behaviors: relevance to autism. Behav. Brain Res. 188, 178–194 (2008).
Nolan, S. O. et al. Deletion of Fmr1 results in sex-specific changes in behavior. Brain Behav. 7, e00800 (2017).
Voikar, V. et al. Automated dissection of permanent effects of hippocampal or prefrontal lesions on performance at spatial, working memory and circadian timing tasks of C57BL/6 mice in IntelliCage. Behav. Brain Res. 352, 8–22 (2018).
Balemans, M. C. et al. Hippocampal dysfunction in the Euchromatin histone methyltransferase 1 heterozygous knockout mouse model for Kleefstra syndrome. Hum. Mol. Genet. 22, 852–866 (2013).
McQuown, S. C. et al. HDAC3 is a critical negative regulator of long-term memory formation. J. Neurosci. 31, 764–774 (2011).
Nuytens, K. et al. Haploinsufficiency of the autism candidate gene Neurobeachin induces autism-like behaviors and affects cellular and molecular processes of synaptic plasticity in mice. Neurobiol. Dis. 51, 144–151 (2013).
Mews, P. et al. Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory. Nature 546, 381–386 (2017).
Peixoto, L. L. et al. Memory acquisition and retrieval impact different epigenetic processes that regulate gene expression. BMC Genomics 16(Suppl 5), S5 (2015).
Alarcon, J. M. et al. Selective modulation of some forms of Schaffer collateral-CA1 synaptic plasticity in mice with a disruption of the CPEB-1gene. Learn. Mem. 11, 318–327 (2004).
Bontekoe, C. J. et al. Knockout mouse model for Fxr2: a model for mental retardation. Hum. Mol. Genet. 11, 487–498 (2002).
Deller, T. et al. Synaptopodin-deficient mice lack a spine apparatus and show deficits in synaptic plasticity. Proc. Natl. Acad. Sci. USA 100, 10494–10499 (2003).
Vigil, F. A., Mizuno, K., Lucchesi, W., Valls-Comamala, V. & Giese, K. P. Prevention of long-term memory loss after retrieval by an endogenous CaMKII inhibitor. Sci. Rep. 7, 4040 (2017).
Zhang, J., Hou, L., Klann, E. & Nelson, D. L. Altered hippocampal synaptic plasticity in the FMR1 gene family knockout mouse models. J. Neurophysiol. 101, 2572–2580 (2009).
Wilkinson, B., Li, J. & Coba, M. P. Synaptic GAP and GEF complexes cluster proteins essential for GTP signaling. Sci. Rep. 7, 5272 (2017).
Zeng, M. et al. Phase transition in postsynaptic densities underlies formation of synaptic complexes and synaptic plasticity. Cell 166, 1163–1175.e12 (2016).
Kim, S. et al. NGL family PSD-95-interacting adhesion molecules regulate excitatory synapse formation. Nat. Neurosci. 9, 1294–1301 (2006).
DeNardo, L. A., de Wit, J., Otto-Hitt, S. & Ghosh, A. NGL-2 regulates input-specific synapse development in CA1 pyramidal neurons. Neuron 76, 762–775 (2012).
Alfieri, A. et al. Synaptic interactome mining reveals p140Cap as a new hub for PSD proteins involved in psychiatric and neurological disorders. Front. Mol. Neurosci. 10, 212 (2017).
Repetto, D. et al. p140Cap regulates memory and synaptic plasticity through Src-mediated and citron-N-mediated actin reorganization. J. Neurosci. 34, 1542–1553 (2014).
Yu, S. E. et al. SET domain-containing protein 5 is required for expression of primordial germ cell specification-associated genes in murine embryonic stem cells. Cell. Biochem. Funct. 35, 247–253 (2017).
Mas-Y-Mas, S. et al. The human mixed lineage leukemia 5 (MLL5), a sequentially and structurally divergent SET domain-containing protein with no intrinsic catalytic activity. PLoS One 11, e0165139 (2016).
Rincon-Arano, H., Halow, J., Delrow, J. J., Parkhurst, S. M. & Groudine, M. UpSET recruits HDAC complexes and restricts chromatin accessibility and acetylation at promoter regions. Cell 151, 1214–1228 (2012).
Perissi, V. et al. TBL1 and TBLR1 phosphorylation on regulated gene promoters overcomes dual CtBP and NCoR/SMRT transcriptional repression checkpoints. Mol. Cell 29, 755–766 (2008).
Gallagher, D. et al. Ankrd11 is a chromatin regulator involved in autism that is essential for neural development. Dev. Cell. 32, 31–42 (2015).
Chen, F. X. et al. PAF1, a molecular regulator of promoter-proximal pausing by RNA polymerase II. Cell 162, 1003–1015 (2015).
Li, J. & Wang, C. Y. TBL1-TBLR1 and beta-catenin recruit each other to Wnt target-gene promoter for transcription activation and oncogenesis. Nat. Cell Biol. 10, 160–169 (2008).
Brandler, W. M. et al. Paternally inherited cis-regulatory structural variants are associated with autism. Science 360, 327–331 (2018).
Grozeva, D. et al. Targeted next-generation sequencing analysis of 1,000 individuals with intellectual disability. Hum. Mutat. 36, 1197–1204 (2015).
Farwell Hagman, K. D. et al. Candidate-gene criteria for clinical reporting: diagnostic exome sequencing identifies altered candidate genes among 8% of patients with undiagnosed diseases. Genet. Med. 19, 224–235 (2017).
Halvardson, J. et al. Mutations in HECW2 are associated with intellectual disability and epilepsy. J. Med. Genet. 53, 697–704 (2016).
Peleg, S. et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science 328, 753–756 (2010).
Kwapis, J. L. et al. Context and auditory fear are differentially regulated by HDAC3 activity in the lateral and basal subnuclei of the amygdala. Neuropsychopharmacology 42, 1284–1294 (2017).
Nott, A. et al. Histone deacetylase 3 associates with MeCP2 to regulate FOXO and social behavior. Nat. Neurosci. 19, 1497–1505 (2016).
Savic, D. et al. CETCh-seq: CRISPR epitope tagging ChIP-seq of DNA-binding proteins. Genome Res. 25, 1581–1589 (2015).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Anders, S., Pyl, P. T. & Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 0550–0558 (2014).
Falcon, S. & Gentleman, R. Using GOstats to test gene lists for GO term association. Bioinformatics 23, 257–258 (2007).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Peixoto, L. et al. How data analysis affects power, reproducibility and biological insight of RNA-seq studies in complex datasets. Nucleic Acids Res. 43, 7664–7674 (2015).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Walter, W., Sánchez-Cabo, F. & Ricote, M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics 31, 2912–2914 (2015).
Gu, Z., Gu, L., Eils, R., Schlesner, M. & Brors, B. Circlize implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812 (2014).
Skene, N. G. & Grant, S. G. Identification of vulnerable cell types in major brain disorders using single cell transcriptomes and expression weighted cell type enrichment. Front. Neurosci. 10, 16 (2016).
Zeisel, A. et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142 (2015).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016). W1.
Dignam, J. D., Lebovitz, R. M. & Roeder, R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 (1983).
Fraser, J. et al. Hierarchical folding and reorganization of chromosomes are linked to transcriptional changes in cellular differentiation. Mol. Syst. Biol. 11, 852 (2015).
Liao, Z., Wan, Y., Thomas, S. N. & Yang, A. J. IsoQuant: a software tool for stable isotope labeling by amino acids in cell culture-based mass spectrometry quantitation. Anal. Chem. 84, 4535–4543 (2012).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016).
Huber, W., von Heydebreck, A., Sültmann, H., Poustka, A. & Vingron, M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18(Suppl 1), S96–S104 (2002).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Jensen, L. J. et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37, D412–D416 (2009).
Goecks, J., Nekrutenko, A. & Taylor, J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11, 10 (2010).
Ramírez, F., Dündar, F., Diehl, S., Grüning, B. A. & Manke, T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Quinlan, A. R. BEDTools: the Swiss-Army tool for genome feature analysis. Curr. Protoc. Bioinformatics 47, 1–34 (2014).
Shen, Y. et al. A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120 (2012).
Ross-Innes, C. S. et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393 (2012).
De Rubeis, S. et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014).
Gräff, J. et al. Epigenetic priming of memory updating during reconsolidation to attenuate remote fear memories. Cell 156, 261–276 (2014).
Holmdahl, R. & Malissen, B. The need for littermate controls. Eur. J. Immunol. 42, 45–47 (2012).
Acknowledgements
We thank V. Stein and M. Döngi for guidance in setting up LTP experiments, P. Jonas for consultancy on LTP data analysis, M. Kutzer for help in statistical analysis of nonparametric datasets, S. Deixler and A. Lepold for management of the animal colony, T. Asenov and his team in the Machine Shop, F. Marr, H.C. Önal, A. Coll, and M. Schunn and the Preclinical Facility team for technical assistance. We thank the EMBL proteomics, flow cytometry, and genomics core facilities for the sample preparation, data generation, and analysis support. We thank M. van Gerven for helping with the SETD5 human mutation study. This work was supported by the Simons Foundation Autism Research Initiative (grant 401299) to G.N. and the DFG (SPP1738 grant NO 1249) to K.-M.N.
Author information
Authors and Affiliations
Contributions
E.D., N.A., J.M., and C.P.D. designed and performed experiments, analyzed data, and prepared figures. X.C. and E.-L.K. performed experiments and data analysis. C.G. supervised data analysis. A.K., K.K., and I.C. performed experiments under the supervision of E.D. and/or J.M. G.N. and K.-M.N. conceived and supervised the study. G.N. wrote the paper together with K.-M.N., E.D., N.A., J.M., and C.D. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Setd5 gene deletion in mice.
a, Schematic diagram for Setd5-floxed allele (top) showing exons 3-6 flanked by loxP sites and the resulting Setd5-knockout allele. b, Typical genotyping results of Setd5+/− and Setd5+/+ mice; the results are shown for single experiment and replication experiments were not performed. c, RT-PCR showing reduction of Setd5 gene expression in the brain of Setd5+/− and Setd5+/+ mice (n(mice)=4 per genotype); Hprt was used as control. Pictures in b and c represent part of the gels; see Supplementary Fig. 14 for full-length picture.
Supplementary Figure 2 Lower survival probability and developmental anomalies in Setd5+/– animals.
a, Doughnut chart indicating that Setd5+/− animals are born at a non-Mendelian ratio (n=585 mice, of which 387 Setd5+/+ and 198 Setd5+/−). b, Kaplan-Meier survival curves showing reduced survival probability of mutant animals (n is 387 for Setd5+/+ and 198 Setd5+/−), which seems to be slightly more pronounced in the females (n is 98 male and 100 female Setd5+/−). c, Increased normalised brain/body weight ratio is observed in P1 male (n=9 controls, n=8 mutants) and female mutant pups (n=6 controls, n=8 mutants) as well as in adult male mutants (>P30, n=10-17) (values normalised to Setd5+/+); *P<0.05, **P<0.01, ***P<0.001; data presented as means and SEM, as well as scatter plots (Supplementary Table 1 contains detailed statistics). d-e, Typical X-ray images of Setd5+/− and Setd5+/+ adult animals; craniofacial maxillary/pre-maxillary abnormalities (d, arrowheads), vertebral fusion abnormalities (d, open arrow) and teeth abnormalities (e, arrowheads) are observed in Setd5+/− mice. The percentage of animals with each of the described phenotype is indicated as doughnut charts (n=13 mice of either genotype; data and pictures (as well as top eye picture in Fig. 1c) taken with permission from International Mouse Phenotyping Consortium, available at http://www.mousephenotype.org/data/genes/MGI:1920145).
Supplementary Figure 3 Generation and gene expression analysis of Setd5-mutant ESC lines.
a, Schematics of CRISPR/Cas9 strategy to generate the Setd5+/− ESC line; two CRISPR guides were used: one targeting the intronic region upstream of exon 3, the other targeting the intronic region downstream of exon 15. The homologous recombination template carries a Red Fluorescent Protein/Puromycin selection cassette flanked by two 1 kb homology arms identical to the regions upstream or downstream of the CRISPR/Cas9 cut sites (green dashed lines). b, Schematic representation of the differentiation process from ESCs to NPCs; LIF: leukemia inhibitory factor; RA: retinoic acid; c, Heatmaps of RNA-seq counts (z-Score) for up-regulated neurodevelopment-associated genes in Setd5+/− at 3 stages of in vitro differentiation. Setd5+/+;tr, CRISPR/Cas9 control lines (tr., transfected). d, Chord plot illustrating the overlap between the in vivo and in vitro DEG datasets. Ribbons show overlaps between sets and ribbon width scales with the number of overlapping genes. Remaining areas represent genes unique for each set. e, Gene ontology (GO) terms enriched in upregulated (top, bars to the right) and downregulated (bottom, bars to the left) differentially expressed genes (adjusted p-value < 0.05) between wild type and Setd5 heterozygous ESCs, EBs and NPCs. Bars show the negative decimal logarithm of the enrichment p-value (shown as negative for downregulated genes). (n=2 wildtype cell lines, 2 CRISPR control lines, and 2 mutant Setd5+/− cell lines, one-sided fisher`s exact test).
Supplementary Figure 4 Setd5 is expressed throughout development and in multiple brain regions.
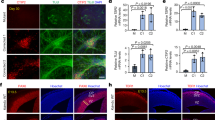
a, Setd5 gene expression in brains of wild type animals at different developmental stages (E16.5, P1, P21, adult) assessed by RT-PCR; Hprt was used as control. b, Setd5-GFP reporter expression in coronal brain sections of E12.5 Setd5GFP embryos and control (Setd5WT), Setd5-GFP is expressed throughout the developing cortex (boxed region and close-up; dashed line: ventricular wall). c, Representative coronal sections of different brain regions of adult control (Setd5WT) and Setd5GFP animals. Broad GFP expression can be observed in several brain regions: cortex (Ctx), hippocampus (Hipp), olfactory bulb (OB) and cerebellum (Crbl). d, Setd5-GFP positive cells co-localize with NeuN-positive neurons, parvalbumin (PV)-positive interneurons and glial fibrillary acidic protein (GFAP)-positive astrocytes in the adult cortex. e, Fluorescent in-situ hybridization (n= one experiment) employing probes against Setd5 exon III-VI and Setd5 exon XIII in adult C57BL/6J sections show nuclear signal and co-localization with Emx1. DAPI nuclear staining (shown in blue) can be seen in (b-e). n=1 Setd5WT and n=3 Setd5GFP. Scale bars: 150 µm in (b); 25 µm in (b, close-up); 50 µm in (c) (Ctx); 200 µm in (c) (Hipp); 25 µm in (c) (OB); 200 µm in (c) (Crbl); 20 µm in (d) (all) and 50 µm in (e). Experiments (a-d) were repeated at least three times, independently, with similar results, using 3 animals per genotype per developmental stage.
Supplementary Figure 5 Normal brain development and adult brain morphology in Setd5+/– mice.
a-f, Representative images (a-c) and quantifications (d-f) of coronal forebrain sections and immunofluorescent stainings at E12.5; dashed line: ventricular wall. a,d, Two hour BrdU incorporation is similar in Setd5+/− and their wild type littermate embryos; b,e, Very low numbers of cleaved Caspase3-positive cells can be found in the developing forebrain of both genotypes (n(mouse embryos)=3 per genotype); c,f, The number of Pax6-positive radial glia in the ventricular zone and Tuj1-positive immature neurons in the developing cortical plate is not changed upon heterozygous loss of Setd5 (n(mouse embryos)=5 per genotype). g, Nissl staining on sagittal sections reveals normal morphology of the adult Setd5+/− brain (n=3 animals per genotype); h-i, Representative images and quantification of the lamination in coronal sections of the somatosensory cortex of Setd5+/+ and Setd5+/− adult brain as assessed by staining for Cux1 positive upper layer and Ctip2 positive lower layer neurons (n=3 animals per genotype); data presented as means and SEM, as well as scatter plots; Scale bars: 25 µm in (a-c); 1 mm in (g); 100 µm in (h), wm: white matter; n.s., not significant, unpaired, two-sided t test, exact p-values and detailed statistics provided in Supplementary Table 1.
Supplementary Figure 6 Further features of Setd5-haploinsufficient animals.
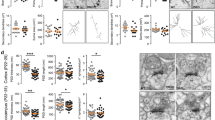
a, Normal exploratory behaviour in the open field. Representative trajectories (a, left), quantifications (a, centre) of velocity, total distance moved, total time spent in the centre (also as measure of anxiety) as well as the distance moved in 5 min bins (a, right) by wild type (grey, Setd5+/+) and mutant (red, Setd5+/−) mice. b, Lack of repetitive behaviours in the open field (b, left) and in marble burying within 30 min (b, right). c, Likewise to the controls, Setd5+/− mice prefer to interact more with a mouse than with an object (normal sociability; left) and favour sniffing a novel mouse over a familiar one (normal preference for social novelty; right) in the three chamber sociability test (a-c, n=25 age and sex-matched littermate pairs of Setd5+/+ and Setd5+/−); d, Performance of male (upper panels) and female (lower panels) Setd5+/− mice in the elevated plus maze (total duration 6 min); Quantification of the total time spent in the closed and open arm as well as arm entries point out natural levels of anxiety, largely identical to the wild type littermates (n=16 male and 10 female age- and sex-matched littermate pairs of wild type and Setd5 heterozygous animals); e, Nest scores showing normal nesting abilities in Setd5+/− males; n=9; f, Isolation-induced ultrasonic vocalisation profile of Setd5+/+ and Setd5+/− male mouse pups, indicating normal number of calls within the first 6 days, followed by failure of mutants to extinguish this type of communication (n=23 male mouse pups per genotype). Data (a-e) presented as means and SEM and scatter plots; boxplots (f) indicate medians (middle line), 25%-75% interquartile range (box) and minimum and maximum data point (whiskers). ***P<0.001; *P<0.05; n.s., not significant (detailed statistics in the Supplementary Table 1); For behavioural tests, in which no inter-genotypic differences were observed, the results of both sexes are presented together, although initially analysed separately. However, for tests in which inter-genotypic differences were discovered, the results are presented for each sex separately.
Supplementary Figure 7 Additional behavioural and electrophysiological features of Setd5+/– mice.
a, Setd5+/− female mice in the Intellicage show more nose pokes per visit (left) but perform significantly less unintentional (no licks) visits with nose pokes (middle) while displaying similar number of licks per visit (right) as compared with the controls (means, SEM and individual data points, n=14 females per genotype). b, Similar freezing behaviour curves during fear memory acquisition (training day, 3 shocks) exclude differences in the pain/sensorial perception between the two genotypes (n=18 adult females per genotype). c, Contextual fear conditioning (CFC) in male mice (strong training, 3 shocks, 5 min total exposure to context, n=15 males per genotype) indicates inability of mutants to extinguish the acquired fear memory when the context becomes a safe environment. Boxplots (c, d) indicate medians (middle line), 25%-75% interquartile range (box) and minimum and maximum data point (whiskers). d, Weak training contextual fear conditioning in male mice (1 shock, 150 s total exposure to context, n=7 males per genotype) indicates that mutants learn the context-shock association after sub-threshold training, and further fail to extinguish it (10 min extinction training in both weak and strong CFC); e, Subthreshold training (3 min) in novel object location memory test induces abnormal memory retention in Setd5 heterozygous males (n=13 males per genotype). Data is presented either as means and SEM, as well as scatter plot; *P<0.05; **P<0.01; ***P<0.001; n.s., not significant; f, Averaged traces (mean ± SEM) indicating the dynamics of fEPSP slope before and up to 5h after LTP induction by high frequency stimulation (HFS, min 0): enhanced late-LTP in the mutants; n=6 (5h), 8 (4h), 10 (3h), 10 (1h) slices for each genotype; one slice per mouse; males, P21-24. g, Similar averaged input-output curves (8 dorsal hippocampus slices from 8 animals per genotype, left panel) and likewise paired-pulse ratios (11 dorsal hippocampus slices from 3 animals per genotype), indicating no difference in the presynaptic function at the hippocampal CA3 to CA1 synapses of P21-24 male Setd5+/+ and Setd5+/- mice. For detailed statistics please refer to Supplementary Table 1.
Supplementary Figure 8 Transcriptional response to CFC in Setd5+/+ and Setd5+–− mice.
a, Volcano plots highlighting Creb1 target genes in the gene expression changes 1h or 3h after conditioning in wild type and mutant animals. The y-axis shows the negative decimal logarithm of the adjusted p-value and the binary logarithm of the fold change is shown on the x-axis; pink, differentially expressed Creb1 target genes; black, other differentially expressed genes; grey, non-significant genes (n(mice)=4 control and 5 mutant animals, EdgeR, likelihood ratio test). b, Example of a DEG cluster showing the same CFC response in Setd5+/+ and Setd5+/− mice (see Methods). Genes listed in the top right corner are early response genes contained in this cluster. Thick lines, average of all genes per cluster; thin lines, single genes; red, Setd5+/−; grey, Setd5+/+; HC, n=5; 1h, n=4; 3h, n=5 per genotype respectively.
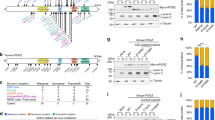
Supplementary Figure 9 SETD5 lacks methyltransferase activity.
a-b, Immunoblots and quantifications evaluating histone H3 methylation states in Setd5 mutant hippocampal and ESC samples. (a) n(mice)=5, N(independent experiments)=3 (b) n(independent cell lines Setd5+/− and Setd5−/−)=2 and 3 respectively, N(independent experiments)=3. No difference in the levels of H3K4-1me, H3K4-2me, H3K4-3me. H3K9-2me, H3K36-3me or H3K36-2me was observed. Data presented as means and SEM (a) or SD (detailed statistics in the Supplementary Table 1) (b). c, Sequence alignment of human SET domain containing proteins. SETD5 and its closest homolog KMT2E (MLL5) are aligned to 13 characterized histone methyltransferase enzymes. Colour scheme: yellow, amino acids involved in SAM binding; blue, amino acids involved in histone H3 recognition. Amino acids in red are the 5 residues in SETD5 that differ the most from the consensus SAM binding motif. The specific histone H3 lysine residues methylated by the characterised SET domains are indicated on the left. d, Schematics of the endogenously C-terminus tagged Setd5. e, PCR genotyping of a successful insertion of the FLAG-HA tag construct (Setd5+/+;HA) compared to a Setd5+/+ control line; FLAG-HA was inserted only on one allele. Experiments were repeated three times, with similar results. f, Western blot results showing the successful tagging of endogenous Setd5. To show that our CRISPR/Cas9 strategy was capable of knocking out Setd5, the two single RNA guides used to generate Setd5−/− (Supplementary Fig. 11a-b) were used on a Setd5+/HA background line and the western blots show complete loss of the Setd5-HA band. Experiments were repeated four times, with similar results. g, Western blot of Setd5-HA immunoprecipitation in non-tagged Setd5+/+ and tagged Setd5+/HA. Membranes blotted for Setd5-HA (left), Tbl1x (right). Arrowheads: Setd5-HA and Tbl1x respectively; asterisk: non-specific band; IN, input: nuclear extract before immunoprecipitation; IP, immunoprecipitation eluate with triple-FLAG peptide; 0.5% of IN and 25% of IP eluate were loaded. Experiment was performed once. h, Volcano plot of proteins identified by quantitative TMT mass spectrometry from Setd5 immunoprecipitation in ESCs and NPCs. Significant proteins (adjusted P value < 0.05 and log2 Fold Change > 0) in red, not significant proteins in grey. Proteins associated with ASD in underlined italics. (n=2 wild type and 2 Setd5 FLAG/HA knock in line, limma statistical analysis).
Supplementary Figure 10 Human SETD5 rescue experiment.
a, Schematics of the localization of de novo SETD5 mutations in ID and ASD patients; each mutation is colour-coded based on the study reporting the de novo mutation; mutations used in this study are highlighted in bold. b-d, Representative example of the bulk enrichment sort and single cell sorting strategy. Shape analysis scatter plots (b). To exclude doublets, live cells are gated on forward scatter/ side scatter (FSC/SSC, left) followed by SSC (middle) and FSC (right). Scatter plots (c) of primary bulk enrichment for Setd5-P2A-GFP and CRISPR/Cas9-T2A-Cerulean3 positive cells (right) compared to WT non-transfected control (left). Scatter plots (d) showing enrichment for Setd5-P2A-GFP cells over the course of the secondary bulk enrichment sort (middle) and tertiary single cell sort (right). Auto-fluorescence was measured to ensure GFP positive cells.
Supplementary Figure 11 Increased H4 acetylation in Setd5 mutants.
a, Schematics of the CRISPR/Cas9 strategy to generate the ΔSET Setd5+/−and Setd5−/− ESC line; two CRISPR guides were used: one targeting the intronic region upstream of exon 7, the other targeting the intronic region downstream of exon 8. b, Schematics of WT and SET domain deleted Setd5 proteins. c, Western blots and quantification of histone post-translational modification in Setd5+/+, Setd5+/− and Setd5−/− ESCs, showing hyperacetylation of histone H4 in Setd5−/−; n(cell lines)= 3 for Setd5+/+ and Setd5−/− and n=2 for Setd5+/−; d-f, Western blot images and quantifications showing H4K8, H4K5, H4K12 and H3 pan acetylation in Setd5+/− CA hippocampal regions dissected from homecage animals (d), 3 hours after CFC training (e) and 1 hour after CFC (f). The band intensity of each acetylated protein residue was normalized to the corresponding total histone level (n(mice)=5 per genotype, N(independent experiments)=3, per time point, respectively). g, H4K8ac dynamics in CA hippocampal samples obtained from homecage and CFC-trained mice showing H4K8 acetylation is significantly higher in samples from homecage (p-value= 0.054) and 3 hours post-training (p-value = 0.009) animals (levels normalized to WT H4K8ac; n(mice)=5 per genotype, n(independent experiments)=3, per timepoint respectively; (two-sided t-test for each timepoint) h, Hdac3 activity assay in brain samples from naïve Setd5+/+ and Setd5+/− mice (n=7). i, Bar plots (means) of de-acetylation activity assay using samples from Hdac3 IP in Setd5+/+ and Setd5−/− ESCs. Dots represent individual technical replicates; n(independent enzymatic reactions)=2 in DMSO and 3 in presence of Trichostatin A (TSA), broad HDAC inhibitor, used as control to show signal specificity towards de-acetylation, data means are shown). Data in c-h presented as means and SEM, **P<0.01; n.s., not significant; detailed statistics in the Supplementary Table 1.
Supplementary Figure 12 ChIP-seq binding profile and differential binding analysis at TSS of Hdac3, Pol II, and H3K27ac.
a-c, Boxplots of input normalised ChIP-seq in Setd5+/+, Setd5+/−, and Setd5−/−. Signal for 4 different genomic regions: Transcription Start Sites (TSS, n=34159) Gene bodies (n=34151), transcription end sites (TES, n=34159), intergenic ESCs specific enhancers (n=4099) for Hdac3 (a), Pol II (b), and H3K27ac (c). d-f, MA plot of differential binding analysis for Setd5+/− (left) and Setd5−/− (right) compared to Setd5+/+ at TSS using peaks of Hdac3 (d), Pol II (e), and H3K27ac (f). EdgeR likelihood ratio test. g, Boxplots of input normalised ChIP-seq RPKM in Setd5+/+, Setd5+/HA, Setd5+/−, and Setd5−/−. Signals of Setd5-HA, Pol II, Hdac3, H3K27ac and H4 pan acetylation for 182 genes that show significant (FDR < 0.05) decrease RNA-Pol II binding in Setd5+/− at TSS. The presence of H3K27ac signal indicates expressed genes. h, Boxplots of ChIP-seq RPKM signal in Setd5+/+, Setd5+/−, and Setd5−/− for 2 different genomic features: TSS having at least one H4 pan acetylation peak (n=3018) and H4 pan acetylation peaks within gene bodies (n=43448 peaks in 9088 gene bodies). TSS is defined as +/- 1.25 kb around gene start position; one-sided Mann–Whitney U test. Boxplots in a, b, c, g, and h represent mean, normalized by input Reads Per Kilobase of genomic regions, per Million mapped reads (RPKM) values of 2 biological replicates from two independent cell lines. Input levels are equal to 1. Outliers, less than 5 or more than 95 percentile of mean RPKM, were removed. The upper, centre, and lower line of the boxplot indicates 75%, 50%, and 25% quantile, respectively. Whiskers extend to the most extreme data point within 1.5-times the interquartile range.
Supplementary Figure 13 Complete ChIP-seq tracks of Meis2 and Neurog1.
ChIP-seq tracks of Setd5-HA in non-tagged (Setd5+/+) and HA-tagged (Setd5+/HA) ESCs and ChIP-seq tracks of Pol II, Hdac3, and H3K27ac in Setd5+/+, Setd5+/−, Setd5−/− ESCs at the Meis2 and Neurog1 gene locus. Presented signals are input-subtracted, merged replicates, Reads per Genomic Content (RPGC). (n=2 independent ChIP biological replicates and n=3 independent input biological replicates for Pol II, Hdac3, and H3K27ac ChIP-seq and n=1 independent input sample for Setd5-HA ChIP-seq with similar results).
Supplementary Figure 14 Full scans of films, gels, and western blots presented in this manuscript.
Dashed orange boxes indicate radioactive film-, gel- and western blot-fragments presented in the respective figure panels.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14
Supplementary Note
Supplementary Note
Supplementary Table 1
Statistics.
Supplementary Table 2
Differentially expressed gene analysis.
Supplementary Table 3
Gene Ontology (GO) term enrichment analysis.
Supplementary Table 4
Differentially expressed gene analysis for CFC samples.
Supplementary Table 5
Cluster identification.
Supplementary Table 6
GO term cluster analysis.
Supplementary Table 7
Setd5 mass spectrometry results.
Supplementary Table 8
PolII ChIP-seq analysis.
Rights and permissions
About this article
Cite this article
Deliu, E., Arecco, N., Morandell, J. et al. Haploinsufficiency of the intellectual disability gene SETD5 disturbs developmental gene expression and cognition. Nat Neurosci 21, 1717–1727 (2018). https://doi.org/10.1038/s41593-018-0266-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-018-0266-2
This article is cited by
-
Transcriptomic dysregulation and autistic-like behaviors in Kmt2c haploinsufficient mice rescued by an LSD1 inhibitor
Molecular Psychiatry (2024)
-
Germline mutations in cancer predisposition genes among pediatric patients with cancer and congenital anomalies
Pediatric Research (2024)
-
SETD5 haploinsufficiency affects mitochondrial compartment in neural cells
Molecular Autism (2023)
-
SETD5 regulates the OGT-catalyzed O-GlcNAcylation of RNA polymerase II, which is involved in the stemness of colorectal cancer cells
Scientific Reports (2023)
-
The Fgf/Erf/NCoR1/2 repressive axis controls trophoblast cell fate
Nature Communications (2023)