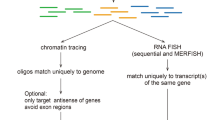
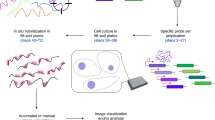
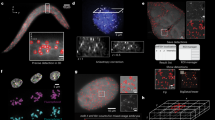
Abstract
Fluorescence in situ hybridization (FISH) allows researchers to visualize the spatial position and quantity of nucleic acids in fixed samples. Recently, considerable progress has been made in developing oligonucleotide (oligo)-based FISH methods that have enabled researchers to study the three-dimensional organization of the genome at super-resolution and visualize the spatial patterns of gene expression for thousands of genes in individual cells. However, there are few existing computational tools to support the bioinformatics workflows necessary to carry out these experiments using oligo FISH probes. Here, we introduce paint server and homology optimization pipeline (PaintSHOP), an interactive platform for the design of oligo FISH experiments. PaintSHOP enables researchers to identify probes for their experimental targets efficiently, to incorporate additional necessary sequences such as primer pairs and to easily generate files documenting library design. PaintSHOP democratizes and standardizes the process of designing complex probe sets for the oligo FISH community.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The original ‘OligoPaints 2012 hg19’ genome-scale probe collection was downloaded from https://oligopaints.hms.harvard.edu/sites/oligopaints.hms.harvard.edu/files/complete-genome-files/hg19.tar.gz. The original OligoMiner hg19 ‘balance’ genome-scale probe collection was downloaded from https://yin.hms.harvard.edu/oligoMiner/probe_seqs/hg19/hg19b.tar.gz. The original OligoMiner hg38 ‘balance’ genome-scale probe collection was downloaded from https://yin.hms.harvard.edu/oligoMiner/probe_seqs/hg38/hg38b.tar.gz. The original ‘Full 40-mer’ iFISH4u hg19 genome-scale probe collection was downloaded from http://ifish4u.org/custom/dbdownload/hg19.gz. The human hg19 genome assembly was downloaded from https://hgdownload.soe.ucsc.edu/goldenPath/hg19/bigZips/hg19.fa.gz. The human hg38 genome assembly was downloaded from https://hgdownload.soe.ucsc.edu/goldenPath/hg38/bigZips/hg38.fa.gz. The mouse mm9 genome assembly was downloaded from https://hgdownload.soe.ucsc.edu/goldenPath/mm9/bigZips/mm9.fa.gz. The mouse mm10 genome assembly was downloaded from https://hgdownload.soe.ucsc.edu/goldenPath/mm10/bigZips/mm10.fa.gz. The C. elegans ce11 genome assembly was downloaded from https://hgdownload.soe.ucsc.edu/goldenPath/ce11/bigZips/ce11.fa.gz. The D. melanogaster dm6 genome assembly was downloaded from https://hgdownload.soe.ucsc.edu/goldenPath/dm6/bigZips/dm6.fa.gz. The zebrafish danRer11 genome assembly was downloaded from https://hgdownload.soe.ucsc.edu/goldenPath/danRer11/bigZips/danRer11.fa.gz. The A. thaliana TAIR10 genome assembly was downloaded from https://www.arabidopsis.org/download_files/Genes/TAIR10_genome_release/TAIR10_chromosome_files/TAIR10_chr_all.fas. The S. cerevisiae sacCer3 genome assembly was downloaded from https://hgdownload.soe.ucsc.edu/goldenPath/sacCer3/bigZips/sacCer3.fa.gz. The rat rn6 genome assembly was downloaded from https://hgdownload.soe.ucsc.edu/goldenPath/rn6/bigZips/rn6.fa.gz. The chicken galGal5 genome assembly was downloaded from https://hgdownload.soe.ucsc.edu/goldenPath/galGal5/bigZips/galGal5.fa.gz. The chicken galGal6 genome assembly was downloaded from https://hgdownload.soe.ucsc.edu/goldenPath/galGal6/bigZips/galGal6.fa.gz. All genome-scale probe collections, primer sequences, bridge sequences, SABER-associated sequences, and transcriptome intersects hosted on paintshop.io are available to download from https://github.com/beliveau-lab/PaintSHOP_resources repository. All repositories are available under the MIT license. Raw and processed microscopy images are available upon request.
Code availability
The source code for the PaintSHOP web application is available as Supplementary Software 1 and at https://github.com/beliveau-lab/PaintSHOP. The source code for the homology optimization pipeline is available as Supplementary Software 2 and at https://github.com/beliveau-lab/PaintSHOP_pipeline.
Change history
03 September 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41592-021-01273-6
References
Pardue, M. L. & Gall, J. G. Molecular hybridization of radioactive DNA to the DNA of cytological preparations. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.64.2.600 (1969).
Rudkin, G. T. & Stollar, B. D. High resolution detection of DNA-RNA hybrids in situ by indirect immunofluorescence. Nature https://doi.org/10.1038/265472a0 (1977).
Bauman, J. G. J., Wiegant, J., Borst, P. & van Duijn, P. A new method for fluorescence microscopical localization of specific DNA sequences by in situ hybridization of fluorochrome-labelled RNA. Exp. Cell Res. https://doi.org/10.1016/0014-4827(80)90087-7 (1980).
Langer-Safer, P. R., Levine, M. & Ward, D. C. Immunological methods for mapping genes on Drosophila polytene chromosomes. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.79.14.4381 (1982).
Moyzis, R. K. et al. A highly conserved repetitive DNA sequence, (TTAGGG)(n), present at the telomeres of human chromosomes. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.85.18.6622 (1988).
Matera, A. G. & Ward, D. C. Oligonucleotide probes for the analysis of specific repetitive dna sequences by fluorescence in situ hybridization. Hum. Mol. Genet. 1, 535–539 (1992).
Dernburg, A. F. et al. Perturbation of nuclear architecture by long-distance chromosome interactions. Cell https://doi.org/10.1016/S0092-8674(00)81240-4 (1996).
Dirks, R. W. et al. Simultaneous detection of different mRNA sequences coding for neuropeptide hormones by double in situ hybridization using FITC- and biotin-labeled oligonucleotides. J. Histochem. Cytochem. https://doi.org/10.1177/38.4.2108203 (1990).
Femino, A. M., Fay, F. S., Fogarty, K. & Singer, R. H. Visualization of single RNA transcripts in situ. Science https://doi.org/10.1126/science.280.5363.585 (1998).
Raj, A., van den Bogaard, P., Rifkin, S. A., van Oudenaarden, A. & Tyagi, S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods https://doi.org/10.1038/nmeth.1253 (2008).
Kosuri, S. & Church, G. M. Large-scale de novo DNA synthesis: technologies and applications. Nat. Methods https://doi.org/10.1038/nmeth.2918 (2014).
Yamada, N. A. et al. Visualization of fine-scale genomic structure by oligonucleotide-based high-resolution FISH. Cytogenet. Genome Res. https://doi.org/10.1159/000322717 (2011).
Boyle, S., Rodesch, M. J., Halvensleben, H. A., Jeddeloh, J. A. & Bickmore, W. A. Fluorescence in situ hybridization with high-complexity repeat-free oligonucleotide probes generated by massively parallel synthesis. Chromosome Res. https://doi.org/10.1007/s10577-011-9245-0 (2011).
Beliveau, B. J. et al. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.1213818110 (2012).
Wang, S. et al. Spatial organization of chromatin domains and compartments in single chromosomes. Science https://doi.org/10.1126/science.aaf8084 (2016).
Bintu, B. et al. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science https://doi.org/10.1126/science.aau1783 (2018).
Cardozo Gizzi, A. M. et al. Microscopy-based chromosome conformation capture enables simultaneous visualization of genome organization and transcription in intact organisms. Mol. Cell https://doi.org/10.1016/j.molcel.2019.01.011 (2019).
Mateo, L. J. et al. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature https://doi.org/10.1038/s41586-019-1035-4 (2019).
Su, J.-H., Zheng, P., Kinrot, S. S., Bintu, B. & Zhuang, X. Genome-scale imaging of the 3D organization and transcriptional activity of chromatin. Cell 182, 1641–1659.e26 (2020).
Takei, Y. et al. Integrated spatial genomics reveals global architecture of single nuclei. Nature https://doi.org/10.1038/s41586-020-03126-2 (2021).
Levesque, M. J. & Raj, A. Single-chromosome transcriptional profiling reveals chromosomal gene expression regulation. Nat. Methods https://doi.org/10.1038/nmeth.2372 (2013).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science https://doi.org/10.1126/science.aaa6090 (2015).
Shah, S. et al. Dynamics and spatial genomics of the nascent transcriptome by intron seqFISH. Cell https://doi.org/10.1016/j.cell.2018.05.035 (2018).
Pernthaler, J., Glöckner, F. O., Schönhuber, W. & Amann, R. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Methods Microbiol. https://doi.org/10.1016/s0580-9517(01)30046-6 (2001).
Yilmaz, L. S., Parnerkar, S. & Noguera, D. R. MathFISH, a web tool that uses thermodynamics-based mathematical models for in silico evaluation of oligonucleotide probes for fluorescence in situ hybridization. Appl. Environ. Microbiol. https://doi.org/10.1128/AEM.01733-10 (2011).
Rogan, P. K., Cazcarro, P. M. & Knoll, J. H. M. Sequence-based design of single-copy genomic DNA probes for fluorescence in situ hybridization. Genome Res. https://doi.org/10.1101/gr.171701 (2001).
Navin, N. et al. PROBER: oligonucleotide FISH probe design software. Bioinformatics https://doi.org/10.1093/bioinformatics/btl273 (2006).
Nedbal, J., Hobson, P. S., Fear, D. J., Heintzmann, R. & Gould, H. J. Comprehensive FISH probe design tool applied to imaging human immunoglobulin class switch recombination. PLoS ONE https://doi.org/10.1371/journal.pone.0051675 (2012).
Bienko, M. et al. A versatile genome-scale PCR-based pipeline for high-definition DNA FISH. Nat. Methods https://doi.org/10.1038/nmeth.2306 (2013).
Baner, J. Parallel gene analysis with allele-specific padlock probes and tag microarrays. Nucleic Acids Res. https://doi.org/10.1093/nar/gng104 (2003).
Stenberg, J., Nilsson, M. & Landegren, U. ProbeMaker: an extensible framework for design of sets of oligonucleotide probes. BMC Bioinf. https://doi.org/10.1186/1471-2105-6-229 (2005).
Rouillard, J. M., Zuker, M. & Gulari, E. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. https://doi.org/10.1093/nar/gkg426 (2003).
Beliveau, B. J. et al. OligoMiner provides a rapid, flexible environment for the design of genome-scale oligonucleotide in situ hybridization probes. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.1714530115 (2018).
Gelali, E. et al. iFISH is a publically available resource enabling versatile DNA FISH to study genome architecture. Nat. Commun. https://doi.org/10.1038/s41467-019-09616-w (2019).
Hu, M. et al. ProbeDealer is a convenient tool for designing probes for highly multiplexed fluorescence in situ hybridization. Sci. Rep. 10, 22031 (2020).
Passaro, M. et al. OligoMinerApp: a web-server application for the design of genome-scale oligonucleotide in situ hybridization probes through the flexible OligoMiner environment. Nucleic Acids Res. 48, W332–W339 (2020).
Kishi, J. Y. et al. SABER amplifies FISH: enhanced multiplexed imaging of RNA and DNA in cells and tissues. Nat. Methods https://doi.org/10.1038/s41592-019-0404-0 (2019).
Köster, J. & Rahmann, S. Snakemake-a scalable bioinformatics workflow engine. Bioinformatics https://doi.org/10.1093/bioinformatics/bts480 (2012).
Casper, J. et al. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. https://doi.org/10.1093/nar/gkx1020 (2018).
Dirks, R. M. & Pierce, N. A. A partition function algorithm for nucleic acid secondary structure including pseudoknots. J. Comput. Chem. https://doi.org/10.1002/jcc.10296 (2003).
Dirks, R. M. & Pierce, N. A. An algorithm for computing nucleic acid base-pairing probabilities including pseudoknots. J. Comput. Chem. https://doi.org/10.1002/jcc.20057 (2004).
Dirks, R. M., Bois, J. S., Schaeffer, J. M., Winfree, E. & Pierce, N. A. Thermodynamic analysis of interacting nucleic acid strands. SIAM Rev. https://doi.org/10.1137/060651100 (2007).
Chen, T. & Guestrin, C. XGBoost: a scalable tree boosting system. in Proc. ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 785–794 (2016); https://doi.org/10.1145/2939672.2939785
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Olson, R. S. et al. Automating biomedical data science through tree-based pipeline optimization. in European Conference on the Applications of Evolutionary Computation 123–137 (Springer, 2016); https://doi.org/10.1007/978-3-319-31204-0_9
Olson, R. S., Bartley, N., Urbanowicz, R. J., & Moore, J. H. Evaluation of a tree-based pipeline optimization tool for automating data science. in Proceedings of the Genetic and Evolutionary Computation Conference 485–492 (2016). https://doi.org/10.1145/2908812.2908918
Smit, A., Hubley, R. & Green, P. RepeatMasker Open-3.0 (Institute for Systems Biology, 1996).
Nir, G. et al. Walking along chromosomes with super-resolution imaging, contact maps, and integrative modeling. PLoS Genet. https://doi.org/10.1371/journal.pgen.1007872 (2018).
Eng, C. H. L. et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+. Nature https://doi.org/10.1038/s41586-019-1049-y (2019).
Kishi, J. Y., Schaus, T. E., Gopalkrishnan, N., Xuan, F. & Yin, P. Programmable autonomous synthesis of single-stranded DNA. Nat. Chem. https://doi.org/10.1038/nchem.2872(2017).
Smit, A., Hubley, R. & Green, P. RepeatMasker Open-4.0. 2013–2015 (Institute for Systems Biology, 2013); http://www.repeatmasker.org
Fornace, M. E., Porubsky, N. J. & Pierce, N. A. A unified dynamic programming framework for the analysis of interacting nucleic acid strands: enhanced models, scalability, and speed. ACS Synth. Biol. 9, 2665–2678 (2020).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics https://doi.org/10.1093/bioinformatics/btq033 (2010).
Anaconda (Anaconda Software Distribution, 2014).
Cock, P. J. A. et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423 (2009).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics https://doi.org/10.1093/bioinformatics/btp352 (2009).
LaFave, M.C. & Burgess, S.M. sam2pairwise v.1.0.0. Zenodo https://doi.org/10.5281/zenodo.11377 (2014).
Marçais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics https://doi.org/10.1093/bioinformatics/btr011 (2011).
Dale, R. K., Pedersen, B. S. & Quinlan, A. R. Pybedtools: a flexible Python library for manipulating genomic datasets and annotations. Bioinformatics 27, 3423–3424 (2011).
Chang, W., Cheng, J., Allaire, J. J., Xie, Y. & McPherson, J. shiny: Web application framework for R. R version 4.0.3 (2019).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2019).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2016).
Sarrab, R.M. et al. Establishment of conditionally immortalized human glomerular mesangial cells in culture, with unique migratory properties. Am. J. Physiol. Renal Physiol. 301, 1131–1138 (2011).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Acknowledgements
We thank G. Nir, D. Shechner, A. Tsue, H. Nguyen, J. Y. Kishi and J. Harke for helpful feedback during the beta testing phase of PaintSHOP, N. Peters and D. Fong for assistance with microscopy, the Genome Sciences IT team for technical assistance and members of the Beliveau laboratory for feedback on the written paper. We also thank S. Lapan and E. West for productive discussions that inspired aspects of this work. This work was supported by a Damon Runyon Dale F. Frey Breakthrough Award (to B.J.B.), the National Institutes of Health (under grant no. 1R35GM137916 to B.J.B.), and the DiaCOMP consortium (under grant no. 19AU3987 to S. Akilesh and B.J.B.). Imaging on the University of Washington W.M. Keck Center Lecia SP8X confocal microscopy was enabled by funding from the NIH (grant no. S10 OD016240). The Nikon CSU-W1 SoRa superresolution microscope was funded by the Howard Hughes Medical Institute. We also thank the Allen Institute for Brain Science founder, P. G. Allen, for his vision, encouragement and support.
Author information
Authors and Affiliations
Contributions
E.A.H., C.K.C. and B.J.B. conceived the study. E.A.H., C.K.C., R.C. and B.J.B. wrote and optimized software code. C.K.C., J.L.C., S. Attar and Y.L. performed validation experiments. E.A.H., C.K.C., J.L.C., S. Akilesh, P.R.N. and B.J.B. conceptualized features of the web application. E.A.H., C.K.C. and B.J.B. wrote the manuscript. All authors edited and approved the paper. S. Akilesh, P.R.N. and B.J.B. supervised the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Methods thanks Zhe Liu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Rita Strack was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8, Tables 1–4 and Notes 1 and 2.
Rights and permissions
About this article
Cite this article
Hershberg, E.A., Camplisson, C.K., Close, J.L. et al. PaintSHOP enables the interactive design of transcriptome- and genome-scale oligonucleotide FISH experiments. Nat Methods 18, 937–944 (2021). https://doi.org/10.1038/s41592-021-01187-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-021-01187-3
This article is cited by
-
Transcription bodies regulate gene expression by sequestering CDK9
Nature Cell Biology (2024)
-
Tigerfish designs oligonucleotide-based in situ hybridization probes targeting intervals of highly repetitive DNA at the scale of genomes
Nature Communications (2024)
-
SGF29 nuclear condensates reinforce cellular aging
Cell Discovery (2023)
-
Research on mobile terminal sketch 3D modeling technology based on interactive design
International Journal on Interactive Design and Manufacturing (IJIDeM) (2023)
-
Interchromosomal interaction of homologous Stat92E alleles regulates transcriptional switch during stem-cell differentiation
Nature Communications (2022)