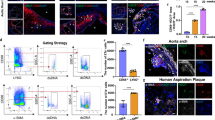
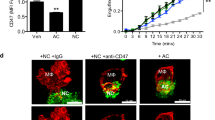
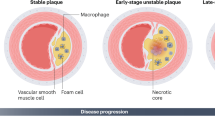
Abstract
The perpetuation of inflammation is an important pathophysiological contributor to the global medical burden. Chronic inflammation is promoted by non-programmed cell death1,2; however, how inflammation is instigated, its cellular and molecular mediators, and its therapeutic value are poorly defined. Here we use mouse models of atherosclerosis—a major underlying cause of mortality worldwide—to demonstrate that extracellular histone H4-mediated membrane lysis of smooth muscle cells (SMCs) triggers arterial tissue damage and inflammation. We show that activated lesional SMCs attract neutrophils, triggering the ejection of neutrophil extracellular traps that contain nuclear proteins. Among them, histone H4 binds to and lyses SMCs, leading to the destabilization of plaques; conversely, the neutralization of histone H4 prevents cell death of SMCs and stabilizes atherosclerotic lesions. Our data identify a form of cell death found at the core of chronic vascular disease that is instigated by leukocytes and can be targeted therapeutically.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
References
Kolb, J. P., Oguin, T. H., III, Oberst, A. & Martinez, J. Programmed cell death and inflammation: winter is coming. Trends Immunol. 38, 705–718 (2017).
Tabas, I. Macrophage death and defective inflammation resolution in atherosclerosis. Nat. Rev. Immunol. 10, 36–46 (2010).
Clarke, M. C. H. et al. Apoptosis of vascular smooth muscle cells induces features of plaque vulnerability in atherosclerosis. Nat. Med. 12, 1075–1080 (2006).
Hartwig, H. et al. Atherosclerotic plaque destabilization in mice: a comparative study. PLoS ONE 10, e0141019 (2015).
Silvestre-Roig, C. et al. Atherosclerotic plaque destabilization: mechanisms, models, and therapeutic strategies. Circ. Res. 114, 214–226 (2014).
Bennett, M. R., Sinha, S. & Owens, G. K. Vascular smooth muscle cells in atherosclerosis. Circ. Res. 118, 692–702 (2016).
Rossaint, J. et al. Synchronized integrin engagement and chemokine activation is crucial in neutrophil extracellular trap-mediated sterile inflammation. Blood 123, 2573–2584 (2014).
Papayannopoulos, V. Neutrophil extracellular traps in immunity and disease. Nat. Rev. Immunol. 18, 134–147 (2018).
Kumar, S. V. et al. Neutrophil extracellular trap-related extracellular histones cause vascular necrosis in severe GN. J. Am. Soc. Nephrol. 26, 2399–2413 (2015).
Tagai, C., Morita, S., Shiraishi, T., Miyaji, K. & Iwamuro, S. Antimicrobial properties of arginine- and lysine-rich histones and involvement of bacterial outer membrane protease T in their differential mode of actions. Peptides 32, 2003–2009 (2011).
Lee, E. Y., Fulan, B. M., Wong, G. C. & Ferguson, A. L. Mapping membrane activity in undiscovered peptide sequence space using machine learning. Proc. Natl Acad. Sci. USA 113, 13588–13593 (2016).
Schmidt, N. W. et al. Criterion for amino acid composition of defensins and antimicrobial peptides based on geometry of membrane destabilization. J. Am. Chem. Soc. 133, 6720–6727 (2011).
Schmidt, N. W. & Wong, G. C. Antimicrobial peptides and induced membrane curvature: geometry, coordination chemistry, and molecular engineering. Curr. Opin. Solid State Mater. Sci. 17, 151–163 (2013).
Franck, G. et al. Roles of PAD4 and NETosis in experimental atherosclerosis and arterial injury: implications for superficial erosion. Circ. Res. 123, 33–42 (2018).
Saffarzadeh, M. et al. Neutrophil extracellular traps directly induce epithelial and endothelial cell death: a predominant role of histones. PLoS ONE 7, e32366 (2012).
Xu, J. et al. Extracellular histones are major mediators of death in sepsis. Nat. Med. 15, 1318–1321 (2009).
Menegazzi, R., Decleva, E. & Dri, P. Killing by neutrophil extracellular traps: fact or folklore? Blood 119, 1214–1216 (2012).
Hasenberg, A. et al. Catchup: a mouse model for imaging-based tracking and modulation of neutrophil granulocytes. Nat. Methods 12, 445–452 (2015).
Dzhagalov, I., St John, A. & He, Y. W. The antiapoptotic protein Mcl-1 is essential for the survival of neutrophils but not macrophages. Blood 109, 1620–1626 (2007).
Virmani, R., Kolodgie, F. D., Burke, A. P., Farb, A. & Schwartz, S. M. Lessons from sudden coronary death: a comprehensive morphological classification scheme for atherosclerotic lesions. Arterioscler. Thromb. Vasc. Biol. 20, 1262–1275 (2000).
Gorelik, R. & Gautreau, A. Quantitative and unbiased analysis of directional persistence in cell migration. Nat. Protoc. 9, 1931–1943 (2014).
Lee, J. et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 (2016).
Wichapong, K. et al. Structure-based design of peptidic inhibitors of the interaction between CC chemokine ligand 5 (CCL5) and human neutrophil peptides 1 (HNP1). J. Med. Chem. 59, 4289–4301 (2016).
van Zundert, G. C. P. et al. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 428, 720–725 (2016).
Alard, J. E. et al. Recruitment of classical monocytes can be inhibited by disturbing heteromers of neutrophil HNP1 and platelet CCL5. Sci. Transl. Med. 7, 317ra196 (2015).
Lee, E. Y., Lee, M. W., Fulan, B. M., Ferguson, A. L. & Wong, G. C. L. What can machine learning do for antimicrobial peptides, and what can antimicrobial peptides do for machine learning? Interface Focus 7, 20160153 (2017).
Lee, E. Y., Wong, G. C. L. & Ferguson, A. L. Machine learning-enabled discovery and design of membrane-active peptides. Bioorg. Med. Chem. 26, 2708–2718 (2018).
McGuffin, L. J., Bryson, K. & Jones, D. T. The PSIPRED protein structure prediction server. Bioinformatics 16, 404–405 (2000).
Schmidt, N. W., Mishra, A., Wang, J., DeGrado, W. F. & Wong, G. C. Influenza virus A M2 protein generates negative Gaussian membrane curvature necessary for budding and scission. J. Am. Chem. Soc. 135, 13710–13719 (2013).
Ilavsky, J. Nika: software for two-dimensional data reduction. J. Appl. Crystallogr. 45, 324–328 (2012).
Hammersley, A. P. FIT2D: a multi-purpose data reduction, analysis and visualization program. J. Appl. Crystallogr. 49, 646–652 (2016).
Necas, D. & Klapetek, P. Gwyddion: an open-source software for SPM data analysis. Cent. Eur. J. Phys. 10, 181–188 (2012).
Chen, S. W. & Pellequer, J. L. DeStripe: frequency-based algorithm for removing stripe noises from AFM images. BMC Struct. Biol. 11, 7 (2011).
Korchev, Y. E. et al. Specialized scanning ion-conductance microscope for imaging of living cells. J. Microsc. 188, 17–23 (1997).
Acknowledgements
The study was supported by the DFG (SFB914 TP B8, SFB1123 TP A6, B5, Z1, SO876/6-1, SO876/11-1, AN372/14-3, AN372/24-1, INST409/97-1FUGG and INST409/150-1FUGG), the EKFS (2016_A118, 2017_A13), the NWO (VIDI project 91712303), the Leducq foundation, and the Vetenskapradet. Funding was also provided by grants SAF2015-65607-R to A.H., BES-2013–065550 to J.M.A., and Severo Ochoa Center of Excellence (award SEV-2015-0505) to CNIC, all from the Ministerio de Ciencia, Innovacion y Universidades. We thank O. Schengel for mouse genotyping and V. Lavilla for generating video animations. This work used the platforms of the Grenoble Instruct-ERIC Center (ISBG: UMS 3518 CNRS-CEA-UGA-EMBL) with support from FRISBI (ANR-10 INSB-05-02) and GRAL (ANR-10-LABX-49-01). E.Y.L. acknowledges support from the Systems and Integrative Biology Training Program (T32GM008185), the Medical Scientist Training Program (T32GM008042), and the Dermatology Scientist Training Program (T32AR071307) at UCLA. E.Y.L. and G.C.L.W. acknowledge an Early Career Research Grant and a Discovery Grant, respectively, from the National Psoriasis Foundation. G.C.L.W. also acknowledges support from NIH R56AI125429-01A1.
Reviewer information
Nature thanks Peter Libby and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
C.S.-R. and Q.B. designed and performed experiments, analysed data, interpreted data and wrote the manuscript. K.W., E.Y.L., J.M.T., N.B., J.W., J.M.A., G.S.S., A.F., A.O.-G., J.M., P.L., R.C., A.S., C. Winter, L.P.-O., C.P., T.S., H.H., J.v.B. and S.G.-R. contributed to data acquisition and analysis; N.P. supervised animal experimentation; F.K., R.T.A.M., H.-J.A., V.O.N., A.H., J.-L.P., G.C.L.W. and G.A.F.N. supervised specific data acquisition and analysis and provided funding; C. Weber provided scientific infrastructure, access to Lyz2creCxcr4flox mice, and contributed to the funding of K.W.; K.A.M. provided Pad4 mice; M.G. provided Ly6gcre mice; T.H. synthetized peptides; and L.M., E.L. and M.D. provided human samples and contributed to data analyses. O.S. conceived and supervised the study, designed experiments and interpreted data, provided funding and wrote the paper. A.H., G.C.L.W. and G.A.F.N. contributed equally to this study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Lesional neutrophils do not correlate with macrophage or endothelial phenotype.
a–f, Methodology used to analyse advanced atherosclerotic lesions. a, b, Representative pictures of H&E staining. a, Intima to media ratio was calculated as the quotient of intima area and media area. Red and black dotted lines delineate intima and media, respectively. b, Necrotic core area was quantified as the percentage of necrotic core area over the intima area. Red and green dotted lines delineate intima and necrotic core, respectively. c, d, Representative pictures of Sirius red staining. c, Sirius red pictures were converted into 8-bit images and segmented by thresholding hence generating binary images. Collagen area was quantified as the percentage of collagen area over the intima area. The black dotted line delineates the intima. d, Fibrous cap thickness was analysed as the average of lengths measurements in the positions overlapping with the lines of a square-shaped grid. Averaged fibrous cap thickness was corrected by the intima area. e, f, Representative pictures of smooth muscle cell actin (SMA, e) and CD68 (f) staining. Immunofluorescence images were segmented by thresholding to convert fluorescence signal into a binary image. SMA/CD68 area was quantified as the percentage of SMA/CD68 area over the intima area. The red dotted line delineates the intima. g–o, Advanced atherosclerotic lesions were generated in the carotid artery of Apoe−/− mice. Pearson correlation between lesional neutrophils and lesion size (n = 28 mice, g), fibrous cap thickness (n = 28 mice, h), collagen area (n = 28 mice, i), macrophage area (CD68+, n = 24 mice, j), pro-inflammatory macrophages (CD68+iNOS+, n = 24 mice, k), anti-inflammatory macrophages (CD68+CD206+, n = 23 mice, l), endothelial cell area (CD31+, n = 25 mice, m), VCAM1 expression on endothelial cells (n = 25 mice, n) and ICAM1 expression on endothelial cells (n = 25 mice, o). Dotted line represents 95% confidence interval. Note that data in g–i and j–o were generated in a different set of lesions due to a shortage of sections from mice displayed in g–i.
Extended Data Fig. 2 Modulation of lesional neutrophil counts alters lesion stability.
a, Models of neutropenia and neutrophilia. b, c, Quantification of lesional neutrophils in indicated treatments. n = 79 sections (ctrl IgG), n = 25 sections (anti-Ly6G), n = 27 sections (PBS), n = 58 sections (AMD3100). Two-sided unpaired t-test. d, e, Lesion volumes of mice with induced neutropenia (d) or induced neutrophilia (e). n = 10 mice (ctrl IgG), n = 9 mice (anti-Ly6G), n = 15 mice (PBS), n = 14 mice (AMD3100). Two-sided unpaired t-test. f–h, Lesion characteristics of mice with antibody-induced neutropenia (anti-Ly6G, n = 10 mice, except in (g) n = 9 mice), genetic neutropenia (Ly6gcreMcl1flox/flox, Mcl1ΔN, n = 16 mice), induced neutrophilia (AMD3100, n = 15 mice) and genetic neutrophilia (Lyz2creCxcr4flox/flox, Cxcr4ΔN, n = 13 mice) compared with respective controls (isotype IgG (n = 10 mice), Ly6gcre (n = 18 mice), vehicle (n = 15 mice) or Lyz2cre (n = 11 mice)), (dashed line). Two-sided unpaired t-test; *P < 0.05; **P < 0.01. Displayed is the quantification of lesion size (f), fibrous cap (FC) thickness (g) and collagen area (h). i, j, Analysis of SMA+MYH11+ (i) and SMA-MYH11+ (j) SMCs in indicated treatments. n = 10 mice (ctrl IgG), n = 9 mice (anti-Ly6G), n = 7 mice (PBS), n = 8 mice (AMD3100). Two-sided unpaired t-test. k–s, Antibody-induced neutropenia generates atherosclerotic lesions with reduced signs of vulnerability in the brachiocephalic artery in Apoe−/− mice fed a HFD for 16 weeks. k, Representative micrographs showing the aortic arch of isotype IgG (ctrl-IgG) and anti-Ly6G-treated mice stained with H&E (top), Sirius red (middle) and antibodies (bottom) to SMA (red), CD68 (green). l, Experimental scheme. m–s, Analyses of lesion characteristics. Displayed are quantification of intima area (m), fibrous cap (FC) thickness (n), necrotic core area (o), collagen area (p), SMC area (SMA+, q), macrophage area (CD68+, r), and plaque vulnerability (s). n = 8 mice (ctrl IgG), n = 9 mice (anti-Ly6G) except for panel (m) n = 8 mice. Two-sided unpaired t-test. t, u, Proliferating SMCs (Ki67+SMA+) in lesions of indicated treatments. n = 8 mice (ctrl IgG), n = 8 mice (anti-Ly6G), n = 12 mice (PBS), n = 13 mice (AMD3100). Two-sided unpaired t-test. v, w, Percentage of SMC area (SMA+, n = 15 sections for 0.51<, n = 25 sections for >0.51, v) and overall vulnerability (n = 10 sections for 0.51<, n = 20 sections >0.51, w) of human atherosclerotic specimens grouped by the number of lesional neutrophils per mm2. Two-sided unpaired t-test. Data are mean ± s.d. BCA, brachiocephalic artery; ctrl, control; SMA, smooth muscle actin; LCA, left carotid artery; LSA, left subclavian artery; MYH11, Myosin heavy chain 11; RCA, right carotid artery.
Extended Data Fig. 3 Activated SMCs induce neutrophil chemotaxis and induce NET-mediated SMC death.
a, Neutrophil displacement in gradient of supernatant obtained from PDGF-BB-activated or resting SMCs (ctrl). n = 20 neutrophils (ctrl), n = 18 neutrophils (PDGF-BB). Two-way ANOVA. P = 1 × 10−15 (ctrl vs PDGF-BB). MSD, mean square displacement. b, Neutrophils transmigrated towards supernatants obtained from PDGF-BB-activated or resting SMCs (ctrl). n = 14 replicates (ctrl), n = 11 replicates (PDGF-BB). Two-sided unpaired t-test. c, d, Multiplex ELISA of indicated growth factors and cytokines (c) and chemokines (d) in cell-free supernatants from SMCs treated with PDGF-BB or vehicle. n = 9 replicates (IL-6, CXCL12), n = 10 replicates (CXCL1, CCL5).Two-sided paired t-test. e, Pearson correlation between neutrophils and intimal CCL7 in mouse advanced atherosclerotic lesions, n = 28 sections. Dotted line represents 95% confidence interval. f, Representative micrographs of mouse atherosclerotic lesions showing SMCs (SMA, green), nuclei (blue), dead cells (TUNEL, red), and NETs (citrullinated histone H3, white). Dashed lines indicate cross-section views. Scale bar, 20 µm. Close-ups represent xz (left) and yz (right) cross-sections. Scale bar, 4 µm. Orange arrows indicate points of interactions between dead SMCs and NETs. g, Micrographs of mouse atherosclerotic lesions showing SMCs (MYH11, white), nuclei (blue), dead cells (TUNEL, red), and MPO (green). Yellow arrows indicate points of interactions between dead SMCs and NETs. Asterisks indicate intact MPO+ cells. h–j, Advanced atherosclerotic lesions in the carotid artery were stained with antibodies to Ly6G, CD68, myeloperoxidase (MPO), and citrullinated H3 (citH3) and counterstained with DAPI. h, Representative images. Scale bar, 50 µm. i, Pie chart showing distribution of macrophage extracellular traps (METs, 1.86%), NETs (80.05%), and extracellular trap DNA (18.09%) based on marker analysis defined underneath, n = 35 sections from 8 mice. j, Extracellular trap DNA structures in carotid artery sections from neutropenic mice (anti-Ly6G, n = 13 sections), mice with intact white blood cell count (vehicle treated, n = 96 sections), or neutrophilic mice (AMD3100, n = 57 sections). Two-sided unpaired t-test. k, Percentage of viable SMCs after exposure to PMA-induced NETs isolated from indicated number of neutrophils. n = 16 biological samples (0, 2.75 × 106 neutrophils), n = 13 biological samples (0.275106, 0.55 × 106, 1.375 × 106, 4.125 × 106 neutrophils), n = 11 biological samples (5.5 × 106 neutrophils). l, Cell death of SMCs incubated with NETs isolated from neutrophils treated with recombinant CCL7. n = 67 fields (−), n = 72 fields (+). Two-sided unpaired t-test, ****P = 0.000002. Data are mean ± s.d. MPO, myeloperoxidase; ND, not detected.
Extended Data Fig. 4 Pharmacological and genetic inhibition of NET release reduces traits of atherosclerotic plaque vulnerability.
a, Experimental scheme. b–g, Quantification of lesion characteristics. Displayed are lesion volumes (b), lesion size (c), fibrous cap (FC) thickness (d), necrotic core area (e), collagen area (f) and macrophage area (CD68+, g). Cl-amidine-treated (Cl-a, n = 9 mice) and Apoe−/−Pad4−/− (Pad4−/−, n = 8 mice) are compared to respective controls (PBS, n = 8 mice), Apoe−/− (Pad4+/+, n = 6 mice). Two-sided unpaired t-tests (PBS vs Cl-a) or two-sided Mann–Whitney tests (Pad4+/+ vs Pad4−/−) were used. h, Quantification of NETs per section of indicated mice. n = 40 sections (PBS), n = 31 sections (Cl-a), n = 35 sections (Apoe−/−), n = 68 sections (Apoe−/−Pad4−/−). Two-sided unpaired t-test. i, j, Analysis of SMA+MYH11+ (i) and SMA−MYH11+(j) SMCs in indicated mice. Two-sided unpaired t-test (PBS vs Cl-a) or two-sided Mann–Whitney test (Pad4+/+ vs Pad4−/−) were used. PBS (n = 8 mice), Cl-amidine (Cl-a, n = 9 mice), Apoe−/− (Pad4+/+, n = 6 mice), Apoe−/−Pad4−/− (Pad4−/−, n = 8 mice). *P < 0.05; **P < 0.01. Data are mean ± s.d.
Extended Data Fig. 5 NET-derived histone H4 induces cell toxicity.
a–c, Analysis of cell death (propidium iodide uptake). a, NETs were pre-incubated with indicated antibodies for 1 h before addition to SMCs. MPO, myeloperoxidase; NE, neutrophil elastase; CG, cathepsin G; PR3, proteinase 3. n = 79 IgG, n = 23 MPO, n = 60 LL37, n = 60 NE, n = 58 CG and n = 60 PR3 fields. One-way ANOVA with Dunnet’s correction. P = 0.105 (MPO), P = 0.219 (LL37), P = 0.270 (NE), P = 0.925 (CG), P = 0.999 (PR3). All conditions were compared against control (ctrl). b, NETs were pre-incubated with inhibitors to myeloperoxidase (MPO), neutrophil elastase (NE), or secretory leukocyte protease (SLP) for 1 h before their addition to SMCs. n = 96 ctrl, n = 35 MPO, n = 58 NE, n = 58 SLP fields. One-way ANOVA with Dunnet’s correction. P = 0.299 (MPO), P = 0.085 (NE), P = 0.978 (SLP). All conditions were compared against control (ctrl). c, SMCs, endothelial cells (ECs) and peritoneal macrophages (PMs) were incubated with recombinant histone H4. Cell death was assessed by PI uptake. n = 36 and n = 36 for SMCs, n = 35 and n = 36 for ECs, n = 47 and n = 39 for PMs. Two-sided unpaired t-test, *P = 0.029; **P = 3.847 × 10−5; ***P = 8.775 × 10−6. d, Representative confocal immunofluorescence of advanced atherosclerotic lesions to visualize DNA (DAPI, blue), neutrophils (Ly6G, red), SMCs (SMA, green), histone H4 (magenta), and citrullinated histone H3 (white). Scale bar, 20 µm. e, Quantification of extranuclear histone H4 per section of indicated treatments. n = 17 ctrl, n = 8 anti-Ly6G, n = 9 AMD3100. One-way ANOVA with Dunnet’s correction, *P = 0.02; **P = 0.0002. Data are mean ± s.d. ctrl, control; SMA, smooth muscle actin.
Extended Data Fig. 6 Neutralization of histone H4 stabilizes atherosclerotic lesions.
a, Experimental scheme. b–i, Quantification of lesion characteristics of the carotid artery. Displayed are lesion volume (b), lesion size (c), fibrous cap (FC) thickness (d), necrotic core area (e), collagen area (f), macrophage area (CD68+, g), SMA+MYH11+ cells (h) and SMA−MYH11+ cells (i). n = 14 mice (ctrl IgG) except for (h, i) n = 11 mice; n = 15 mice (anti-histone H4) except for (h, i) n = 12 mice. Two-sided unpaired t-test. j–p, Quantification of lesion characteristics on the brachiocephalic artery. Displayed are lesion size (j), fibrous cap (FC) thickness (k), necrotic core area (l), collagen area (m), SMCs (SMA+, n), macrophages (CD68+, o) and overall vulnerability (p). Two-sided unpaired t-test, n = 12 mice (ctrl IgG) or 10 mice (anti-histone H4). Data are mean ± s.d.
Extended Data Fig. 7 NET-derived histone H4 interaction with cell membranes is surface charge dependent and induces a lytic cell death.
a, SMCs were pre-incubated with indicated inhibitors before NET treatment. Cell death was assessed by PI uptake. n = 24 fields, except TLR4, n = 23 fields. One-way ANOVA with Dunnet’s correction, P = 0.729 (TLR1/2), P = 0.999 (TLR3), P = 0.995 (TLR4). All conditions were compared against control (ctrl). b, Representative high-resolution confocal microscopy images were used to visualize cell membrane (lectin, white), histone H4 (magenta) and DNA (DAPI, cyan) in a SMC (S) and neutrophil (N) co-culture. Dashed lines indicate cross-section views represented in Fig. 3j. c, Confocal immunofluorescence micrograph to visualize cell membrane (lectin, white), histone H4 (magenta) and DNA (DAPI, cyan). The dashed line indicates the cross-section view represented in Fig. 3k. d, e, SMCs were incubated with NETs. d, Extracellular ATP. n = 3 biological replicates (crtl), n = 6 biological replicates (NETs). Two-sided Mann–Whitney test. e, Flow cytometry analysis of cell size. n = 9 biological replicates. Two-sided unpaired t-test. f, g, SMCs were incubated with histone H4. f, Extracellular ATP. n = 5 biological replicates. Two-sided Mann–Whitney test. g, Time-lapse microscopy images were used to measure SMC area before and after incubation with histone H4. n = 9 cells. Two-sided paired t-test. h, Analysis of the ζ potential of SMCs incubated with oleylamine or cholesterol sulfate (chl sulfate). n = 9 biological replicates (ctrl), n = 8 biological replicates (oleylamine), n = 6 biological replicates (chl sulfate). Two-sided Mann–Whitney test. i, j, SMCs were incubated with recombinant histone H4 after pre-incubation with oleylamine or cholesterol sulfate (chl sulfate). i, Confocal microscopy was used to detect histone H4 and plasma cell membrane (phalloidin). Peptide–membrane interaction quantified as the ratio of histone H4-fragment signal and plasma membrane area. n = 10 cells (ctrl), n = 20 cells (histone H4, −), n = 20 cells (oleylamine), n = 25 cells (chl sulfate). One-way ANOVA with Dunnet’s correction. **P = 0.007; ***P = 0.0001 vs ctrl. j, Quantification of PI incorporation. n = 54 fields, n = 8 fields, n = 10 fields, n = 34 fields, n = 21 fields and n = 19 fields for each condition represented. One-way ANOVA with Tukey’s correction, *P = 0.001; **P = 0.004. Data are mean ± s.d.
Extended Data Fig. 8 Membrane pore-forming activity of histone H4.
a, Scanning electron micrographs of SMCs incubated with recombinant histone H4 or vehicle. b, Machine learning screen of full-length sequence histone H4 predicts potent membrane activity at the N terminus (residues 1–24 highlighted in blue). c, SAXS data demonstrates that N-terminal domain of histone H4 induces negative Gaussian curvature (NGC) in cell membranes at the indicated peptide:lipid (P/L) ratios. The histone H4 N terminus was incubated with indicated membrane compositions and the resulting structures were measured with SAXS. The peptide induced Pn3m cubic phases, which are rich in NGC, and are indicative of membrane permeation. d, SMCs were incubated with biotinylated histone H4 fragments (1–24: N terminus; 25–68: α-helix; 69–102: C terminus). Confocal microscopy was used to detect histone H4 fragments and plasma cell membrane. Peptide–membrane interaction was quantified as the ratio of histone H4 fragment signal and plasma membrane area. n = 44 cells (1–24), n = 28 cells (25–68), n = 33 cells (69–102). One-way ANOVA with Tukey’s correction; *P = 0.049; **P = 4 × 10−14. e, PI incorporation in SMCs treated with histone H4 fragments or the full-length protein. n = 19 fields (ctrl), n = 24 fields (histone H4), n = 24 fields (1–24), n = 21 fields (25–68), n = 19 fields (69–102).One-way ANOVA with Dunnet’s correction; *P = 0.005; **P = 0.0001 vs control. f, Histone H4 was pre-incubated with HIPe or vehicle and added to SMCs. Confocal microscopy was used to visualize interaction of histone H4 (green) with plasma cell membrane (phalloidin, red). n = 20 cells (ctrl), n = 17 cells (histone H4), n = 15 cells (histone H4+HIPe). One-way ANOVA with Tukey’s correction; *P = 9.243 × 10−7; **P = 6.239 × 10−9. Scale bar, 20 µm. g, Atomic force microscopy studies of lipid membranes treated with the indicated histone H4:HIPe ratio. Scale bar, 1 µm. Membrane disruption was quantified as membrane roughness. n = 13 membranes (ctrl), n = 3 membranes (1:0), n = 3 (1:1). Kruskal–Wallis test with Dunn’s correction. h, Live scanning ion conductance microscopy of SMCs. Images represent the plasma membrane before and after incubation with histone H4 and HIPe. i, PI incorporation in SMCs treated with recombinant histone H4 in the presence or absence of HIPe. n = 33 fields (ctrl), n = 12 fields (histone H4), n = 11 fields (histone H4 + HIPe). One-way ANOVA with Tukey’s correction; *P = 0.001; **P = 8.844 × 10−6. Data are mean ± s.d.
Extended Data Fig. 9 Disruption of histone H4–cell membrane interaction prevents pore formation and stabilizes atherosclerotic lesions.
a, Experimental scheme. b–i, Lesion characteristics in the carotid artery. Displayed are lesion volume (b), lesion size (c), fibrous cap (FC) thickness (d), necrotic core area (e), collagen area (f), macrophage area (CD68+, g), SMA+MYH11+ (h) and SMA−MYH11+ (i) SMCs. n = 12 mice (vehicle), n = 11 mice (HIPe) except for (h) n = 9 mice and (i) n = 10 mice. Two-sided unpaired t-test. j, Experimental scheme. k–q, Assessment of lesion characteristics. Displayed is quantification lesion size (k), fibrous cap (FC) thickness (l), necrotic core area (m), collagen area (n), SMC area (SMA+, o), macrophage area (p) and overall vulnerability (q). n = 4 mice (vehicle), n = 4 mice (sHIPe). Two-sided Mann–Whitney test. r–x, Assessment of lesion characteristics in the brachiocephalic artery of mice treated as described in a. Displayed is quantification lesion size (r), fibrous cap (FC) thickness (s), necrotic core area (t), collagen area (u), SMCs (SMA+, v), macrophages (CD68+, w) and overall vulnerability (x). n = 11 mice (vehicle), n = 11 mice (HIPe), except for (s) Vehicle n = 10. Two-sided unpaired t-test, y, Number of lesional neutrophils per section of vehicle- (n = 82 cells) or HIPe- (n = 83 cells) treated mice. Two-sided unpaired t-test. z, Mice were treated with TNF and received either vehicle control or HIPe. Neutrophils were quantified in the peritoneum (left, n = 4 (vehicle), n = 5 (HIPe)), the bone marrow (middle, n = 5 (vehicle), n = 5 (HIPe)), and the blood (right, n = 5 (vehicle), n = 5 (HIPe)). aa, Scheme summarizing the cytotoxic activity of neutrophils during atherosclerotic plaque vulnerability. The interaction of lesional SMCs with neutrophils induces NETosis (1) which acts as a carrier of cytotoxic histone H4 (2). The cationicity of the N-terminal domain of histone H4 induces plasma membrane bending and pore formation leading to SMC lysis (3). SMC death contributes to fibrous cap thinning and increased plaque instability. Data are mean ± s.d.
Supplementary information
41586_2019_1167_MOESM1_ESM.pdf
Supplementary Figure Supplementary Figure 1: Source data Histone H4-membrane interaction western blot. SMCs treated with recombinant histone H4 were labelled with sulfo-NHS-SS-biotin. Western blot of non-purified lysates (SMCs) and plasma membrane fractions (SMCs avidin-purified) with indicated antibodies. Red line delineates cropped area as shown in Figure 4l.
41586_2019_1167_MOESM3_ESM.mov
Video 1: Histone H4 generates membrane permeability. Time-lapse microscopy of calcein-loaded smooth muscle cells treated with recombinant histone H4 (50 µg/mL) in presence of propidium iodide. Images were taken every 30 seconds over a period of 15 min and show calcein (green) dispersion and propidium iodide (red) entry.
41586_2019_1167_MOESM4_ESM.avi
Video 2: Histone H4 binds to the plasma membrane of smooth muscle cells. Smooth muscle cell treated with histone H4 (50 µg/mL, 5 min, 4°C). In motion overview of 3D reconstructed image showing a smooth muscle cell stained with lectin (plasma membrane, white), histone H4 (magenta) and DAPI (nucleus, cyan).
Source data
Rights and permissions
About this article
Cite this article
Silvestre-Roig, C., Braster, Q., Wichapong, K. et al. Externalized histone H4 orchestrates chronic inflammation by inducing lytic cell death. Nature 569, 236–240 (2019). https://doi.org/10.1038/s41586-019-1167-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1167-6
This article is cited by
-
Diagnostic value of the creatine kinase-MB/creatine kinase and neutrophil/lymphocyte ratios in acute myocardial infarction
Journal of Cardiothoracic Surgery (2024)
-
What is the actual relationship between neutrophil extracellular traps and COVID-19 severity? A longitudinal study
Respiratory Research (2024)
-
Vascular smooth muscle cells in response to cholesterol crystals modulates inflammatory cytokines release and promotes neutrophil extracellular trap formation
Molecular Medicine (2024)
-
Neutrophil extracellular traps: a catalyst for atherosclerosis
Molecular and Cellular Biochemistry (2024)
-
Neutrophil extracellular traps in acute coronary syndrome
Journal of Inflammation (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.