Abstract
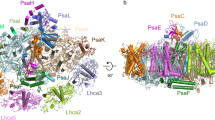
Photosystem I (PSI) possesses a variable supramolecular organization among different photosynthetic organisms to adapt to different light environments. Mosses are evolutionary intermediates that diverged from aquatic green algae and evolved into land plants. The moss Physcomitrium patens (P. patens) has a light-harvesting complex (LHC) superfamily more diverse than those of green algae and higher plants. Here, we solved the structure of a PSI–LHCI–LHCII–Lhcb9 supercomplex from P. patens at 2.68 Å resolution using cryo-electron microscopy. This supercomplex contains one PSI–LHCI, one phosphorylated LHCII trimer, one moss-specific LHC protein, Lhcb9, and one additional LHCI belt with four Lhca subunits. The complete structure of PsaO was observed in the PSI core. One Lhcbm2 in the LHCII trimer interacts with PSI core through its phosphorylated N terminus, and Lhcb9 mediates assembly of the whole supercomplex. The complicated pigment arrangement provided important information for possible energy-transfer pathways from the peripheral antennae to the PSI core.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The cryo-EM density map and atomic model for the PSI–LHCI–LHCII–Lhcb9 supercomplex structure at 2.68 Å resolution have been deposited in the Electron Microscopy Data Bank and the Protein Data Bank (EMD ID code 33401 and PDB ID code 7XQP). The data that support the findings of this study are available from the corresponding authors upon request. Source data are provided with this paper.
References
Nelson, N. & Junge, W. Structure and energy transfer in photosystems of oxygenic photosynthesis. Annu. Rev. Biochem. 84, 659–683 (2015).
Croce, R. & van Amerongen, H. Natural strategies for photosynthetic light harvesting. Nat. Chem. Biol. 10, 492–501 (2014).
Croce, R. & van Amerongen, H. Light harvesting in oxygenic photosynthesis: structural biology meets spectroscopy. Science 369, eaay2058 (2020).
Cao, P., Pan, X., Su, X., Liu, Z. & Li, M. Assembly of eukaryotic photosystem II with diverse light-harvesting antennas. Curr. Opin. Struct. Biol. 63, 49–57 (2020).
Suga, M. & Shen, J.-R. Structural variations of photosystem I–antenna supercomplex in response to adaptations to different light environments. Curr. Opin. Struct. Biol. 63, 10–17 (2020).
Jordan, P. et al. Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature 411, 909–917 (2001).
Malavath, T., Caspy, I., Netzer-El, S. Y., Klaiman, D. & Nelson, N. Structure and function of wild-type and subunit-depleted photosystem I in Synechocystis. Biochim. Biophys. Acta 1859, 645–654 (2018).
Kato, K. et al. Structure of a cyanobacterial photosystem I tetramer revealed by cryo-electron microscopy. Nat. Commun. 10, 4929 (2019).
Zheng, L. et al. Structural and functional insights into the tetrameric photosystem I from heterocyst-forming cyanobacteria. Nat. Plants 5, 1087–1097 (2019).
Chen, M. et al. Distinct structural modulation of photosystem I and lipid environment stabilizes its tetrameric assembly. Nat. Plants 6, 314–320 (2020).
Semchonok, D. A. et al. Cryo-EM structure of a tetrameric photosystem I from Chroococcidiopsis TS-821, a thermophilic, unicellular, non-heterocyst-forming cyanobacterium. Plant Commun. 3, 100248 (2021).
Toporik, H. K., Li, J., Williams, D., Chiu, P. L. & Mazor, Y. The structure of the stress-induced photosystem I–IsiA antenna supercomplex. Nat. Struct. Mol. Biol. 26, 443–449 (2019).
Cao, P. et al. Structural basis for energy and electron transfer of the photosystem I–IsiA–flavodoxin supercomplex. Nat. Plants 6, 167–176 (2020).
Akita, F. et al. Structure of a cyanobacterial photosystem I surrounded by octadecameric IsiA antenna proteins. Commun. Biol. 3, 232 (2020).
Xu, C. et al. A unique photosystem I reaction center from a chlorophyll d-containing cyanobacterium Acaryochloris marina. J. Integr. Plant Biol. 63, 1740–1752 (2021).
Hamaguchi, T. et al. Structure of the far-red light utilizing photosystem I of Acaryochloris marina. Nat. Commun. 12, 2333 (2021).
Kato, K. et al. Structural basis for the absence of low-energy chlorophylls in a photosystem I trimer from Gloeobacter violaceus. eLife 11, e73990 (2022).
Pi, X. et al. Unique organization of photosystem I–light-harvesting supercomplex revealed by cryo-EM from a red alga. Proc. Natl Acad. Sci. USA 115, 4423–4428 (2018).
Qin, X. et al. Structure of a green algal photosystem I in complex with a large number of light-harvesting complex I subunits. Nat. Plants 5, 263–272 (2019).
Su, X. et al. Antenna arrangement and energy transfer pathways of a green algal photosystem-I–LHCI supercomplex. Nat. Plants 5, 273–281 (2019).
Suga, M., Ozawa, S. I., Yoshida-Motomura, K., Akita, F. & Takahashi, Y. Structure of the green algal photosystem I supercomplex with a decameric light-harvesting complex I. Nat. Plants 5, 626–636 (2019).
Perez-Boerema, A., Klaiman, D., Caspy, I., Netzer-El, S. Y. & Nelson, N. Structure of a minimal photosystem I from the green alga Dunaliella salina. Nat. Plants 6, 321–327 (2020).
Caspy, I., Malavath, T., Klaiman, D., Fadeeva, M. & Nelson, N. Structure and energy transfer pathways of the Dunaliella salina photosystem I supercomplex. Biochim. Biophys. Acta Bioenerg. 1861, 148253 (2020).
Caspy, I. et al. Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii. Nat. Plants 7, 1314–1322 (2021).
Nagao, R. et al. Structural basis for assembly and function of a diatom photosystem I–light-harvesting supercomplex. Nat. Commun. 11, 2481 (2020).
Xu, C. et al. Structural basis for energy transfer in a huge diatom PSI–FCPI supercomplex. Nat. Commun. 11, 5081 (2020).
Qin, X., Suga, M., Kuang, T. & Shen, J.-R. Structural basis for energy transfer pathways in the plant PSI–LHCI supercomplex. Science 348, 989–995 (2015).
Mazor, Y., Borovikova, A., Caspy, I. & Nelson, N. Structure of the plant photosystem I supercomplex at 2.6 Å resolution. Nat. Plants 3, e17014 (2017).
Wang, J. et al. Structure of plant photosystem I–light harvesting complex I supercomplex at 2.4 Å resolution. J. Integr. Plant Biol. 63, 1367–1381 (2021).
Ruban, A. V. & Johnson, M. P. Dynamics of higher plant photosystem cross-section associated with state transitions. Photosynth. Res. 99, 173–183 (2009).
Minagawa, J. & Tokutsu, R. Dynamic regulation of photosynthesis in Chlamydomonas reinhardtii. Plant J. 82, 413–428 (2015).
Goldschmidt-Clermont, M. & Bassi, R. Sharing light between two photosystems: mechanism of state transitions. Curr. Opin. Plant Biol. 25, 71–78 (2015).
Allen, J. F., Bennett, J., Steinback, K. E. & Arntzen, C. J. Chloroplast protein phosphorylation couples plastoquinone redox state to distribution of excitation energy between photosystems. Nature 291, 25–29 (1981).
Rochaix, J. D. et al. Protein kinases and phosphatases involved in the acclimation of the photosynthetic apparatus to a changing light environment. Philos. Trans. R. Soc. B 367, 3466–3474 (2012).
Rochaix, J. D. Regulation and dynamics of the light-harvesting system. Annu. Rev. Plant Biol. 65, 287–309 (2014).
Rantala, M., Rantala, S. & Aro, E. M. Composition, phosphorylation and dynamic organization of photosynthetic protein complexes in plant thylakoid membrane. Photochem. Photobiol. Sci. 19, 604–619 (2020).
Pan, X. W. et al. Structure of the maize photosystem I supercomplex with light-harvesting complexes I and II. Science 360, 1109–1113 (2018).
Huang, Z. H. et al. Structure of photosystem I–LHCI–LHCII from the green alga Chlamydomonas reinhardtii in State 2. Nat. Commun. 12, 1100 (2021).
Pan, X. et al. Structural basis of LhcbM5-mediated state transitions in green algae. Nat. Plants 7, 1119–1131 (2021).
Rensing, S. A. et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69 (2008).
Alboresi, A., Caffarri, S., Nogue, F., Bassi, R. & Morosinotto, T. In silico and biochemical analysis of Physcomitrella patens photosynthetic antenna: identification of subunits which evolved upon land adaptation. PLoS ONE 3, e2033 (2008).
Iwai, M. & Yokono, M. Light-harvesting antenna complexes in the moss Physcomitrella patens: implications for the evolutionary transition from green algae to land plants. Curr. Opin. Plant Biol. 37, 94–101 (2017).
Iwai, M. et al. Light-harvesting complex Lhcb9 confers a green alga-type photosystem I supercomplex to the moss Physcomitrella patens. Nat. Plants 1, 14008 (2015).
Iwai, M., Grob, P., Iavarone, A. T., Nogales, E. & Niyogi, K. K. A unique supramolecular organization of photosystem I in the moss Physcomitrella patens. Nat. Plants 4, 904–909 (2018).
Pinnola, A. et al. LHCB9-dependent photosystem I megacomplex induced under low light in Physcomitrella patens. Nat. Plants 4, 910–919 (2018).
Yan, Q. et al. Antenna arrangement and energy-transfer pathways of PSI–LHCI from the moss Physcomitrella patens. Cell Discov. 7, 10 (2021).
Gorski, C. et al. The structure of the Physcomitrium patens photosystem I reveals a unique Lhca2 paralogue replacing Lhca4. Nat. Plants 8, 307–316 (2022).
Alboresi, A., Gerotto, C., Cazzaniga, S., Bassi, R. & Morosinotto, T. A red-shifted antenna protein associated with photosystem II in Physcomitrella patens. J. Biol. Chem. 286, 28978–28987 (2011).
Morosinotto, T., Breton, J., Bassi, R. & Croce, R. The nature of a chlorophyll ligand in Lhca proteins determines the far-red fluorescence emission typical of photosystem I. J. Biol. Chem. 278, 49223–49229 (2003).
Romero, E. et al. The origin of the low-energy form of photosystem I light-harvesting complex Lhca4: mixing of the lowest exciton with a charge-transfer state. Biophys. J. 96, L35–L37 (2009).
Su, X. et al. Structure and assembly mechanism of plant C2S2M2-type PSII–LHCII supercomplex. Science 357, 815–820 (2017).
Graham, L. E., Kim, E., Arancibia-Avila, P., Graham, J. M. & Wilcox, L. W. Evolutionary and ecophysiological significance of sugar utilization by the peat moss Sphagnum compactum (Sphagnaceae) and the common charophycean associates Cylindrocystis brebissonii and Mougeotia sp. (Zygnemataceae). Am. J. Bot. 97, 1485–1491 (2010).
Schween, G., Hohe, A., Koprivova, A. & Reski, R. Effects of nutrients, cell density and culture techniques on protoplast regeneration and early protonema development in a moss, Physcomitrella patens. J. Plant Physiol. 160, 209–212 (2003).
Porra, R. J., Thompson, W. A. & Kriedemann, P. E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394 (1989).
Ikeuchi, M. & Inoue, Y. A new 4.8-kDa polypeptide intrinsic to the PSII reaction center, as revealed by modified SDS–PAGE with improved resolution of low-molecular-weight proteins. Plant Cell Physiol. 29, 1233–1239 (1988).
Angeler, D. G. & Schagerl, M. Distribution of the xanthophyll loroxanthin in selected members of the Chlamydomonadales and Volvocales (Chlorophyta). Phyton 37, 119–132 (1997).
Manaresi, E. et al. Chemiluminescence western blot assay for the detection of immunity against parvovirus B19 VP1 and VP2 linear epitopes using a videocamera based luminograph. J. Virol. Methods 81, 91–99 (1999).
Schulenberg, B., Goodman, T. N., Aggeler, R., Capaldi, R. A. & Patton, W. F. Characterization of dynamic and steady-state protein phosphorylation using a fluorescent phosphoprotein gel stain and mass spectrometry. Electrophoresis 25, 2526–2532 (2004).
Schorb, M., Haberbosch, I., Hagen, W., Schwab, Y. & Mastronarde, D. Software tools for automated transmission electron microscopy. Nat. Methods 16, 471–477 (2019).
Zheng, S. Q. et al. Motioncor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. CryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Grigorieff, N. & Harrison, S. C. Near-atomic resolution reconstructions of icosahedral viruses from electron cryo-microscopy. Curr. Opin. Struct. Biol. 21, 265–273 (2011).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Pettersen, E. F. et al. UCSF chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of coot. Acta Crystallogr. D 66, 486–501 (2010).
Liu, Z. et al. Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature 428, 287–292 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Acknowledgements
We thank W. Tang and Y. Yin from the Institute of Botany, Chinese Academy of Sciences (CAS) for instrumental supports in sample preparation, fluorescence measurement and HPLC analysis; the Centre of Cryo-Electron Microscopy, Zhejiang University for their technical assistance on cryo-EM data collection; C. Ma from the Protein Facility, School of Medicine, Zhejiang University for providing the platform for sample purification; and X. Meng at the Centre of Biomedical Analysis, Tsinghua University for protein MS analysis. The project was funded by the National Key R&D Programme of China (2022YFC1803400, 2022YFA0911900, 2020YFA0907600, 2018YFA0507700, 2019YFA0906300, 2021YFA1300403), CAS Project for Young Scientists in Basic Research (YSBR-004), the Strategic Priority Research Programme of CAS (XDA27050402), CAS Interdisciplinary Innovation Team (JCTD-2020-06), Youth Innovation Promotion Association of CAS (2020081), the Fundamental Research Funds for the Central Universities (2018XZZX001-13) and KAKENHI JP22H04916.
Author information
Authors and Affiliations
Contributions
G.H., J.-R.S. and X.Z. conceived the project. S.Z., Q.Y. and G.H. performed the sample preparation and characterization. K.T. collected the cryo-EM data. X.Z. processed the cryo-EM data and reconstructed the cryo-EM map. S.Z., X.L., L.S. and W.W. built the structural model and refined the structure. S.Z., G.H., X.Z. and J.-R.S. analysed the structure. S.Z., K.T., G.H., X.Z. and J.-R.S. jointly wrote the manuscript. All authors discussed and commented on the results and the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Roberto Bassi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 SDS-PAGE analysis and identification of the phosphorylation state of protein subunits in the PSI-LHCI-LHCII-Lhcb9 supercomplex of P. patens.
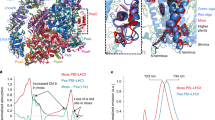
a, SDS-PAGE analysis of marker (Lane 1), thylakoid (Lane 2), purified PSI-LHCI (Lane 3) and purified PSI-LHCI-LHCII-Lhcb9 (Lane 4). Each band was identified based on the mass spectrometry and proteomics analysis and indicated. b, Western-blotting analysis of thylakoid membrane, PSI-LHCI and PSI-LHCI-LHCII-Lhcb9 supercomplex using the antibodies against Lhcb9. c, Western-blotting analysis of the phosphoproteins in thylakoid membrane, PSI-LHCI and PSI-LHCI-LHCII-Lhcb9 supercomplex using a phosphothreonine antibody. d, Detection of the phosphoproteins by Pro-Q Diamond staining. The polypeptide samples were separated by SDS-PAGE and stained with Coomassie blue (left) or with Pro-Q Diamond to detect the presence of phosphoproteins (right). The amounts of the samples loaded onto each wells 2-4 were 10, 5 and 5 μg of Chls in a, b, c and d, respectively. Data presented in this figure is repeated more than three times, and all resulted in the same results.
Extended Data Fig. 2 Data collection and image processing.
a, b, Representative images of negative staining micrograph (a) and cryo-EM micrograph (b) of the PSI-LHCI-LHCII-Lhcb9 supercomplex from P. patens. The negative staining image was taken more than five times, and all resulted in similar particles. The cryo-EM micrograph was taken for 4,000 images, which were used in the subsequent structural analysis. c, Flow chart of the cryo-EM data processing for the Pp PSI-LHCI-LHCII-Lhcb9 supercomplex. d, Gold standard FSC curves of the final 3D reconstruction of the Pp PSI-LHCI-LHCII-Lhcb9 supercomplex. e, Local resolution distributions of the Pp PSI-LHCI-LHCII-Lhcb9 generated with cryoSPARC.
Extended Data Fig. 3 Cryo-EM density maps of protein subunits and representative cofactors in the PSI-LHCI-LHCII-Lhcb9 supercomplex of P. patens.
The PSI core and LHC antenna are shown as cartoon and coloured differently. Pigments (Chl a, Chl b, Bcr, Lut, Neo and Vio), lipids (1,2-dipalmitoyl-phosphatidyl-glycerol (PG), 1,2-distearoyl-monoylgalactosyl-diglyceride (MGDG), digalactosyl diacylglycerol (DGDG)) and other ligands (phylloquinone (PQN), iron/sulfur cluster (SF4) and n-dodecyl-α-D-maltoside (LMU)) are represented by sticks. The cryo-EM density maps of each subunit and cofactors are depicted in grey meshes.
Extended Data Fig. 4 Sequence alignment of Lhca1 isoforms and the assignment of the protein subunit in the structure of Pp PSI-LHCI-LHCII-Lhcb9.
a, Sequence alignment of three Lhca1 isoforms of P. patens. b, Assignment of Lhca1.2 into the structure of Pp PSI-LHCI-LHCII-Lhcb9 by comparison of the map features of characteristic residues and the corresponding sequences. Lhca1.2 contains residues Asn136, Pro170, and His233 which fit well with the density map, which excludes the existence of the other two Lhca1.1 and Lhca1.3 proteins. The red triangles show characteristic residues used for identification. Sequences are taken from Uniprot as follows: Lhca1.1[PHYPADRAFT_139567 - Chlorophyll a-b binding protein, chloroplastic - Physcomitrium patens (Spreading-leaved earth moss) - PHYPADRAFT_139567 gene & protein (uniprot.org)]. Lhca1.2[PHYPADRAFT_218647 - Chlorophyll a-b binding protein, chloroplastic - Physcomitrium patens (Spreading-leaved earth moss) - PHYPADRAFT_218647 gene & protein (uniprot.org)]. Lhca1.3[PHYPADRAFT_146725 - Chlorophyll a-b binding protein, chloroplastic - Physcomitrium patens (Spreading-leaved earth moss) - PHYPADRAFT_146725 gene & protein (uniprot.org)].
Extended Data Fig. 5 Sequence alignment of Lhca2 isoforms and the assignment of the protein subunit in the structure of Pp PSI-LHCI-LHCII-Lhcb9.
a, Sequence alignment of five Lhca2 isoforms of P. patens. b, Assignment of Lhca2.2 by comparison of the map features of characteristic residues and the corresponding sequences. Lhca2.2 contains residues Phe69, Ala148, Pro150, Asn174, Val237, Leu252, Gly259 and Phe264 which fit well with the density map, which excludes the existence of all other Lhca2 isoform proteins. c, Assignment of Lhca2.5 by comparison of the map features of characteristic residues and the corresponding sequence. Lhca2.5 contains residues Leu69, Asn101, Tyr139, Lys140, Gln150, Arg174, Phe237, Phe252, Tyr259and Leu264 which fit well with the density map, which excludes the existence of all other Lhca2 isoform proteins. The red triangles show characteristic residues used for identification. Sequences are taken from Uniprot as follows: Lhca2.1[PHYPADRAFT_178694 - Chlorophyll a-b binding protein, chloroplastic - Physcomitrium patens (Spreading-leaved earth moss) - PHYPADRAFT_178694 gene & protein (uniprot.org)]. Lhca2.2[PHYPADRAFT_151155 - Chlorophyll a-b binding protein, chloroplastic - Physcomitrium patens (Spreading-leaved earth moss) - PHYPADRAFT_151155 gene & protein (uniprot.org)]. Lhca2.3[PHYPA_025494 - Chlorophyll a-b binding protein, chloroplastic - Physcomitrium patens (Spreading-leaved earth moss) - PHYPA_025494 gene & protein (uniprot.org)]. Lhca2.4[PHYPADRAFT_63624 - Chlorophyll a-b binding protein, chloroplastic - Physcomitrium patens (Spreading-leaved earth moss) - PHYPADRAFT_63624 gene & protein (uniprot.org)]. Lhca2.5[PHYPA_014361 - Chlorophyll a-b binding protein, chloroplastic - Physcomitrium patens (Spreading-leaved earth moss) - PHYPA_014361 gene & protein (uniprot.org)].
Extended Data Fig. 6 Sequence alignment of Lhca3 isoforms and the assignment of the protein subunit in the structure of Pp PSI-LHCI-LHCII-Lhcb9.
a, Sequence alignment of Lhca3.1 and Lhca3.3 isoforms of P. patens. b, Assignment of Lhca3.1 by comparison of the map features of characteristic residues and the corresponding sequences. Lhca3.1contains unique residues Gln264, Val271, and Thr286 which fit well with the density map, which excludes the existence of the Lhca3.3 isoform proteins. The red triangles show characteristic residues used for identification. Sequences are taken from Uniprot as follows: Lhca3.1[PHYPA_009318 - Chlorophyll a-b binding protein, chloroplastic - Physcomitrium patens (Spreading-leaved earth moss) - PHYPA_009318 gene & protein (uniprot.org)]. Lhca3.3[PHYPA_021044 - Chlorophyll a-b binding protein, chloroplastic - Physcomitrium patens (Spreading-leaved earth moss) - PHYPA_021044 gene & protein (uniprot.org)].
Extended Data Fig. 7 Sequence alignment of Lhca2.5 and Lhca5 and the identification of Lhca2.5 subunit in the Pp PSI-LHCI-LHCII-Lhcb9 structure.
a, Sequence alignment of Lhca2.5 and Lhca5 from P. patens. Sequences are taken from the following Uniprot. b-h, Identification of the Lhca2.5 subunit in LHCI belt I of Pp PSI-LHCI-LHCII-Lhcb9 by comparison of the cryo-EM density map (b) features of characteristic amino acid residues from Lhca2.5 (green) and Lhca5 (yellow). The possibility of Lhca5 as a Lhca in the structure of the Pp PSI-LHCI-LHCII-Lhcb9 is excluded on the basis of structural features of characteristic residues (c-h), respectively. The sequences used are: Lhca2.5[PHYPA_014361 - Chlorophyll a-b binding protein, chloroplastic - Physcomitrium patens (Spreading-leaved earth moss) - PHYPA_014361 gene & protein (uniprot.org)]. Lhca5[PHYPA_006083 - Chlorophyll a-b binding protein, chloroplastic - Physcomitrium patens (Spreading-leaved earth moss) - PHYPA_006083 gene & protein (uniprot.org)].
Extended Data Fig. 8 Structural comparisons of Lhcb9 from P. patens with Lhcbm5 from C. reinhardtii and Lhcb6 (CP24) from P. sativum.
a, Structural comparisons of Lhcb9 from Pp PSI-LHCI-LHCII-Lhcb9 (PDB code: 7XQP, green) with Lhcbm5 from PSI-LHCI (PDB code: 7DZ7, cyan) of C. reinhardtii and Lhcb6 of PSI-LHCI (PDB code: 5XNL, blue violet) of P. sativum. The Lhcbs are shown with ribbon models, whereas the pigments and lipids are shown as sticks and coloured as that of the protein subunits respectively. Different regions between the three homologues subunits are highlighted by black dashed boxes. The phytol chains of all Chls were omitted for clarity.
Extended Data Fig. 9 Structural characterization of LHCII in Pp PSI-LHCI-LHCII-Lhcb9.
a, Cryo-EM density of LHCII trimer (Lhcbm2a, Lhcbm2b, and Lhcbm2c), viewed from the stromal side. b, Cryo-EM density map of Lhcbm2c, PsaH, PsaL, PsaO. c, Structural comparisons of Lhcbm2 from Pp PSI-LHCI-LHCII-Lhcb9 (PDB code: 7XQP, crimson) with Lhcbm1 from PSI-LHCI-LHCII (PDB code: 7DZ7, cyan) of C. reinhardtii and Lhcb2 from PSI-LHCI (PDB code: 5XNL, blue violet) of Z. mays. The Lhcbs are shown with ribbon models, whereas the pigments are shown as sticks and coloured as that of the protein subunits, respectively. Regions with difference between the three homologues subunits are highlighted by black dashed boxes. The phytol chains of all Chls were omitted for clarity. The map is shown as a semitransparent surface.
Extended Data Fig. 10 Sequence alignment of Lhcbm isoforms and the assignment of the LHCII trimer protein subunits in the structure of Pp PSI-LHCI-LHCII-Lhcb9.
a, Sequence alignment of Lhcbm isoforms of P. patens. b, Assignment of Lhcbm2 by comparison of the map features of characteristic residues and the corresponding sequences. Lhcbm2 possesses residues Thr70, Ser163, Ser121and Pro166 which fit well with the density map, which excludes the existence of all other Lhcbm proteins. The red triangles show characteristic residues used for identification. Sequences are taken from Uniprot as follows: Lhcbm1[PHYPA_022864 - Chlorophyll a-b binding protein, chloroplastic - Physcomitrium patens (Spreading-leaved earth moss) - PHYPA_022864 gene & protein (uniprot.org)]. Lhcbm2[PHYPA_025699 - Chlorophyll a-b binding protein, chloroplastic - Physcomitrium patens (Spreading-leaved earth moss) - PHYPA_025699 gene & protein (uniprot.org)]. Lhcbm5[PHYPA_017103 - Chlorophyll a-b binding protein, chloroplastic - Physcomitrium patens (Spreading-leaved earth moss) - PHYPA_017103 gene & protein (uniprot.org)]. Lhcbm6[PHYPA_030328 - Chlorophyll a-b binding protein, chloroplastic - Physcomitrium patens (Spreading-leaved earth moss) - PHYPA_030328 gene & protein (uniprot.org)]. Lhcbm7[PHYPA_007358 - Chlorophyll a-b binding protein, chloroplastic - Physcomitrium patens (Spreading-leaved earth moss) - PHYPA_007358 gene & protein (uniprot.org)]. Lhcbm8[PHYPA_003988 - Chlorophyll a-b binding protein, chloroplastic - Physcomitrium patens (Spreading-leaved earth moss) - PHYPA_003988 gene & protein (uniprot.org)]. Lhcbm9[PHYPA_003692 - Chlorophyll a-b binding protein, chloroplastic - Physcomitrium patens (Spreading-leaved earth moss) - PHYPA_003692 gene & protein (uniprot.org)]. Lhcbm11[PHYPA_003701 - Chlorophyll a-b binding protein, chloroplastic - Physcomitrium patens (Spreading-leaved earth moss) - PHYPA_003701 gene & protein (uniprot.org)]. Lhcbm13[PHYPA_006353 - Chlorophyll a-b binding protein, chloroplastic - Physcomitrium patens (Spreading-leaved earth moss) - PHYPA_006353 gene & protein (uniprot.org)]. Lhcbm14[PHYPA_003990 - Chlorophyll a-b binding protein, chloroplastic - Physcomitrium patens (Spreading-leaved earth moss) - PHYPA_003990 gene & protein (uniprot.org)].
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2 and Fig. 1.
Supplementary Table 1
Mass spectroscopic data.
Source data
Source Data Fig. 1
Unprocessed gels and western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, S., Tang, K., Yan, Q. et al. Structural insights into a unique PSI–LHCI–LHCII–Lhcb9 supercomplex from moss Physcomitrium patens. Nat. Plants 9, 832–846 (2023). https://doi.org/10.1038/s41477-023-01401-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-023-01401-4