Abstract
Mycobacterium chelonae is a rare cause of chronic disseminated cutaneous infections in immunocompromised patients. Multidrug-resistant M. chelonae infections present a challenge for treatment, and prolonged antimicrobial courses lead to significant toxicities and further antimicrobial resistance. We report a case of refractory cutaneous disseminated M. chelonae infection in a patient with seronegative arthritis on immunotherapy with tofacitinib that was treated with combination antimicrobial, surgical, and single bacteriophage therapy with excellent clinical response. The patient developed neutralizing antibodies against the bacteriophage but continues to have stable improvement of disease with negative biopsies and no evidence of bacterial resistance to the phage.
Similar content being viewed by others
Introduction
Mycobacterium chelonae, a rapidly growing nontuberculous mycobacterium, is a rare cause of chronic infections in immunocompromised hosts1,2. This organism is ubiquitous in the environment and most frequently manifests clinically as localized skin or soft tissue infection or disseminated cutaneous disease1,3,4. Nosocomial infections can occur following procedures such as plastic surgery or exposure to contaminated equipment or substances, both in healthcare settings and community settings such as tattoo parlors5,6,7,8. Catheter-related bloodstream infections, septic arthritis, and osteomyelitis are also reported, while pulmonary disease is less common1. Disseminated cutaneous disease occurs most commonly in immunosuppressed patients including those who have undergone solid organ transplantation or with autoimmune disorders receiving treatment with corticosteroids or immunotherapy1,9,10. Similar to other mycobacterial species such as Mycobacterium abscessus, widely known for extensive antimicrobial resistance, M. chelonae has also been demonstrated to be multidrug-resistant1,10,11,12. Here we report the clinical course of a man with refractory disseminated cutaneous M. chelonae infection who was treated with a single bacteriophage (Muddy) in combination with antimicrobial therapy with excellent clinical response. To our knowledge this is the first case of M. chelonae infection treated with bacteriophage therapy.
Results
Clinical course
A 56-year-old man presented to dermatology clinic in January of 2020 with new nodular lesions on his left upper extremity as well as weight loss, night sweats, myalgias, and fatigue. He had a history of stage II chronic kidney disease, myxomatous mitral valve prolapse requiring mitral valve repair in 2014, and seronegative arthritis involving his bilateral hands, wrists, knees, and ankles diagnosed in 2019, as well as peripheral neuropathy of unclear etiology that was diagnosed simultaneously with his arthritis. Over the year preceding his presentation, he was treated with multiple immunologic therapies for his arthritis including methotrexate, etanercept, adalimumab, and most recently tofacitinib since November 2019 with minimal response. He also received multiple courses of prednisone up to 40 mg per day. At the time of presentation, he was on treatment with 10 mg per day of oral prednisone.
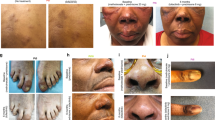
In the clinic, he described several months of waxing and waning nodular lesions on the left upper extremity with intermittent spontaneous drainage (Fig. 1A–B). He denied any known trauma or penetrating injury to that region. A biopsy was performed with mycobacterial cultures that grew M. chelonae. Pathology demonstrated abundant Mycobacteria on staining with dense infiltrates of admixed inflammatory cells and necrosis. The organism susceptibilities are presented in Table 1. The patient’s course of antimicrobial treatment has been characterized by a series of severe toxicities resulting in modification of treatment (Fig. 2). The patient started treatment with oral azithromycin, oral trimethoprim-sulfamethoxazole (TMP/SMX) and intravenous tobramycin on January 23, 2020. His course was complicated by hospitalization in late February with worsening symptoms thought to be due to immune reconstitution inflammatory syndrome in the setting of tapering prednisone. Further immunologic therapy including tofacitinib was held. He also developed multiple medication toxicities including renal injury and ototoxicity due to tobramycin and gastrointestinal symptoms due to TMP/SMX, leading to a period of azithromycin monotherapy and the development of subsequent resistance. Mycobacterial isolator cultures were negative, but in June 2020 he developed new lesions in the periumbilical subcutaneous tissue as well as on the right lower extremity indicative of disseminated cutaneous infection. A positron emission tomography/computed tomography (PET/CT) scan showed no visceral sites of disease (Fig. 1C), and a transthoracic echocardiogram was negative for valvular vegetations. Workup was also notable for the presence of hypogammaglobulinemia and a variant of uncertain significance in the NLRP12 gene on whole exome sequencing, which has been associated with immune dysregulation and familial autoinflammatory disorders. He had no other evidence of underlying immunodeficiencies. He was treated with several new antimicrobial medications over the subsequent months including tedizolid, omadacycline, clofazimine, bedaquiline, imipenem, TMP/SMX, and meropenem-vaborbactam, that were limited by toxicities or emergence of resistance as shown in Table 1 and Fig. 2. He had persistent skin lesions with positive mycobacterial cultures and positive mycobacterial staining on pathology in May, August, and October of 2020 despite efforts to optimize his antimicrobial regimens. In December 2020, he developed severe left wrist pain and was found to have septic arthritis of the left wrist. He underwent a left wrist synovectomy with irrigation and debridement on December 14, 2020 as well as debulking of his most active skin lesions with pathology that demonstrated necrotizing granulomas, though cultures remained negative. He underwent reinduction with a 5-drug regimen of meropenem-vaborbactam, TMP/SMX, omadacycline, clofazimine, and bedaquiline for one month and was then transitioned to a 3-drug regimen of omadacycline, clofazimine, and bedaquiline. He continued on this regimen until February 2021, when he developed worsening nodular and fluctuant lesions, and repeat biopsy demonstrated florid dermal abscesses with numerous Mycobacteria on Fite and acid-fast bacilli (AFB) stains. Given his refractory infection, the patient was identified as a potential candidate for bacteriophage therapy.
A Left upper extremity with multiple large erythematous, fluctuant to nodular lesions ultimately diagnosed as disseminated cutaneous Mycobacterium chelonae infection. B Images of the left upper extremity lesions prior to (Dec 2020) and following (August 2021) addition of bacteriophage therapy. C PET/CT prior to (March 2021) and following (August 2021) addition of bacteriophage therapy.
Identification of suitable therapeutic phages
The M. chelonae isolate from October 2020 (designated strain GD153) was used to screen for potentially therapeutic phages. GD153 was tested for sensitivity to a panel of ~20 Mycobacterium smegmatis phages that have previously shown potential for activity against M. abscessus infections (Fig. 3A), as well as nine lytically growing phages induced from lysogenic M. abscessus clinical isolates (Fig. 3B)13. M. chelonae GD153 does not grow at 37 °C and all phage assays were performed at 30 °C. A single phage—Muddy (and its host range variant Muddy_HRMGD04) – efficiently infects GD153 (Fig. 3A). Phage Muddy is lytic and efficiently kills M. chelonae GD153 with no evident survival after challenging 4 ×108 cells with phage at a multiplicity of infection of ten (Fig. 3C). Muddy also kills M. chelonae GD153 well over a broad range of bacteria and phage concentrations (Fig. 3D). Muddy has a siphoviral morphology, grows well to high titer, is stable (maintains titer), efficiently kills M. tuberculosis, and has been used therapeutically to treat M. abscessus infections14,15. Muddy was grown to high titer on M. smegmatis, highly purified, and shown to be endotoxin-free and sterile, as described previously14,15. Regulatory and institutional approval for intravenous mycobacteriophage administration was obtained from the Massachusetts General Hospital/Brigham and Women’s Hospital institutional review board and the U. S. Food and Drug Administration via a single patient expanded access application.
A Ten-fold serial dilutions of each phage (left) were spotted onto top agar overlays of M. smegmatis mc2155 and M. chelonae GD153. Phages: 1, BPs∆33HTH_HRM10; 2, BPs∆33HTH_HRMGD03; 3, Muddy; 4, Muddy_HRMGD04; 5, Itos; 6, Adephagia∆41∆43; 7, ZoeJ∆45; 8, Fionnbharth∆45∆47; 9, Elmo_HRMmc2155; 10, Isca_cpm; 11, Fred313cpm_∆33; 12, CrimD∆41-43; 13- BPs∆33HTH_HRM10_REM1; 14, Faith1∆38-40; 15, Faith1∆38-40_HRMGD69; 16, MissWhite; 17, MissWhite-D29_Hybrid1, 18, Maco6; 19, TM4; 20, Wildcat; 21, D29; and 22, D29_HRMGD40. B As for panel A but using M. abscessus lytically induced prophages plated on M. chelonae GD153 and control strain M. abscessus GD40. Phages: 1, phiGD20-1; 2, phiGD22-1; 3, phiGD23-1; 4, phiGD21-1; 5, phiGD34-2; 6, phiGD89-1; 7, phiGD57-1; 8, phiGD17-1; 9, phiGD24-3. C Killing of M. chelonae GD153 by Muddy following infection in liquid culture (moi, 10); -Muddy, no phage control. D Efficient killing of M. chelonae GD153 over ranges of bacterial and phage concentrations. All rows contain 10-fold serial dilutions of bacterial culture. Top row (–), no phage control; rows 2-9, 10-fold serial diltions, with 1010 PFU in row 2. E ELISA responses for anti-Muddy IgG antibodies in sera from 3 days, 17 days or 16 weeks after phage treatment initiation. Negative controls (ctrl) for uncoated wells are also shown. n = 2 technical replicates are shown. Data points are the average of two independent sets of serum dilutions and measurements, with error bars ± one standard deviation. F Half-maximal IgG titers derived from ELISA curve fits in panel D. Duplicate measurements are shown as points; bar height is the mean titer with error bars±one standard deviation. Source data for panels E and F are provided as a Source Data file. G To test for antibody neutralization, Muddy was incubated with either plasma (p) or serum (s) from 2 days, 3 days, 17 days or 16 weeks after the start of phage administration for 2 h (top) or 24 h (bottom) and then 10-fold serial dilutions were spotted onto top agar overlays of M. smegmatis mc2155. Plates were incubated at 37 °C for 48 h.
Phage administration and patient response
In the month preceding initiation of bacteriophage therapy, the patient continued on treatment with omadacycline, bedaquiline, and TMP/SMX. He underwent a second debulking surgery of his largest skin lesions on June 10, 2021 with pathology that demonstrated abundant necrotizing granulomas that stained positive for AFB. He started intravenous bacteriophage therapy twice daily at a dose of 109 plaque-forming units (PFUs) per dose of Muddy on June 15, 2021 in conjunction with his prior antimycobacterial therapy. He reported subjective flushing after each dose that lasted up to an hour, but denied fevers, chills, rigors, respiratory symptoms, gastrointestinal symptoms, or neurologic symptoms. He remained hemodynamically stable with stable laboratory markers. Two days after initiation, he experienced chills and nausea two hours after infusion of the bacteriophage without other associated symptoms or vital sign changes. These findings resolved without intervention. He was monitored with examination and weekly laboratory testing including a basic metabolic panel, liver function tests, and complete blood count with differential following discharge from the hospital on June 18, 2021, where he was noted to be doing well without any reported adverse effects and stable laboratory markers. His skin lesions improved significantly over the first two weeks of therapy and continued to show steady improvement over the subsequent months with decreased inflammation and nodularity (Fig. 1B). Small subcutaneous nodules persisted, so repeat biopsies were performed in August and November 2021, and showed no evidence of granulomas or AFB on histopathology and tissue cultures were negative. A repeat PET/CT in September 2021 demonstrated significant interval improvement of size and intensity of FDG uptake in previously seen cutaneous and subcutaneous soft tissue lesions (Fig. 1C).
Serum from days 3 and 17, and week 16 after initiation of phage administration was tested for the presence of antibodies against Muddy (Fig. 3E, F). The day 3 sample showed a weak but specific IgG response that presumably pre-dated phage administration, but at both day 17 and week 16 post-phage initiation there was a robust IgG-mediated response, comparable to that reported in a prior patient15. The antibody response at 17-days was mildly neutralizing but more potently neutralizing at 16 weeks (Fig. 3G). These antibody responses did not negate the overall clinical improvement of the patient. Given his marked improvement and lack of adverse effects, the patient has continued on intravenous bacteriophage therapy. He has had intermittent flares of his seronegative arthritis and additional immunosuppressive therapy is planned with concern that alteration in the host immune system could impact control of infection, bacteriophage and antibiotics have been continued. He will continue on combination therapy for treatment of M. chelonae until at least 4 – 6 weeks after initiation of additional immunosuppressive treatments with the plan to sequentially discontinue the bacteriophage followed by antibiotics thereafter. The patient consented to the publication of this information.
Discussion
Bacteriophages are organisms that are ubiquitous in the environment with the ability to infect and kill bacterial hosts16,17,18. Bacteriophage therapy using a cocktail of multiple phage isolates has been recently successfully used for the treatment of resistant bacterial infections including multidrug-resistant nontuberculous mycobacterial infections in conjunction with traditional antibiotic therapy14,15,19,20. Widespread use of bacteriophage therapy is limited by logistic and regulatory challenges as well as the development of bacterial resistance or neutralizing antibodies that may reduce host response to the bacteriophage15,21,22.
Here, we describe the novel treatment of a patient with disseminated cutaneous Mycobacterium chelonae infection with a single bacteriophage in conjunction with antibiotic and surgical management. While he had previously received extensive courses of antimicrobials as well as prior surgical debridement in December 2020, his skin lesions and biopsies remained culture positive with evidence of necrotizing granulomas and abundant mycobacteria on staining. Following initiation of bacteriophage therapy, the skin lesions significantly improved both on examination and radiographically on PET/CT scan. Furthermore, two biopsies at 2 and 5 months post-treatment have demonstrated no evidence of granulomas or AFB on histopathology and tissue cultures have remained negative. The patient has had no adverse events related to his bacteriophage therapy and has successfully administered this therapy intravenously at home for > 6 months.
A potential major limitation of bacteriophage therapy is the development of phage resistance, which can potentially be countered using an appropriately-designed phage cocktail14,19. Here, we were limited to a single phage as no other phages tested were highly active against the patient’s strain of M. chelonae. Although resistance to Muddy is likely to occur, it was not detected in vitro, consistent with the infrequency of phage resistance in M. abscessus isolates13. Moreover, phage resistance in vivo leading to loss of treatment efficacy was not observed. These observations suggest that phage resistance of NTM pathogens may not be the impediment encountered with other pathogens.
A second barrier to the successful treatment of bacterial infections with phage therapy is the complex interaction between the host immune system and the bacteriophage15,22. The development of neutralizing antibodies has previously been reported, though the clinical consequences remain unclear23. In one case by Dedrick at al., the emergence of neutralizing antibodies correlated temporally with treatment failure, however other reports have shown favorable outcomes resulting from bacteriophage therapy despite the development of anti-phage antibodies15,22. In our case, the patient has maintained stably improved disease and negative microbiologic and histopathologic studies despite the identification of a neutralizing antibody response to the phage. It is plausible that the bacteriophage acted to rapidly decrease the burden of infection, leading to improved control with ongoing antimicrobial therapy that had previously been insufficient. Another consideration is that phage replication became self-sustaining at the sites of infection, such that administration subsequent to the onset of the neutralizing response had little effect and was unnecessary.
To our knowledge, this is the first case of a human M. chelonae infection treated with a bacteriophage, and the first case of bacteriophage treatment with a single phage for a mycobacterial infection. Bacteriophage therapy is a promising therapeutic option for multi-drug resistant infections, though improved understanding of safety, the factors driving the development of bacterial resistance, and the clinical significance of antibody-mediated phage neutralization is vital to advance this therapeutic option for patients.
Methods
Bacterial strains
M. smegmatis mc2155 was grown as previously described.24 M. chelonae GD153 and M. abscessus GD40 were grown in Middlebrook 7H9 media with OADC and 1 mM CaCl2, shaking, at 30 °C for 10-12 days and 37 °C for 4-5 days, respectively13. For plaque assays, M. chelonae and M. abscessus cultures were sonicated briefly in a cup-horn sonicator (Q-sonica 500) at 30% amplitude with 15 s on and 10 s off until visibly dispersed 14.
Phage susceptibility screening
A standard plaque assay was used to determine phage susceptibility of GD153, spotting 10-fold serial dilutions of each phage onto top agar overlays containing 500 µl of saturated cultures of each bacterial isolate.24 The most concentrated phage spot was typically 108 -109 PFU/ml; TM4 was 106 PFU/ml. Phages used were primarily those shown previously to infect one or more strain of M. abscessus or which were recovered as induced prophages from M. abscessus clinical isolates13,15. Efficiencies of plaquing (EOP) were determined by comparing phage titers on M. smegmatis mc2155, M. chelonae GD153 and M. abscessus GD40 isolates, as described previously.24
Therapeutic mycobacteriophage preparation
Phage Muddy was grown on M. smegmatis mc2155 using solid media using a top layer containing 0.35% agar. After phage growth, the whole top layer was collected and centrifuged to remove debris and cells. The clarified lysate (3 × 1011 PFU ml−1) was filtered through a 0.22 μm filter and the phage collected by centrifugation at 100,000 x g for one hour. The phage pellet was resuspended in phage buffer (68 mM NaCl, 10 mM Tris HCl pH 7.5, 10 mM MgSO4, 10 mM CaCl2) and cesium chloride (CsCl) was added to a density of 1.5 g cm−3 (4.1 M), and subjected to equilibrium density gradient centrifugation at 132,600 x g for 16 h. The visible phage band was collected (1-3 ml), the volume increased with CsCl (1.5 g cm−3) and similarly centrifuged. The visible phage band was collected and stored at 4 °C; this yielded ~2 ml of purified phage with a titer of ~1013 PFU ml−1. One ml of Muddy was dialyzed against 1 L of Ringers Solution (Oxoid) four times for a minimum of 4 hours each, and endotoxin shown to be below the level of detection using the EndoZyme II (Hyglos GmbH) assay. USP71 sterility testing was completed by Accugen. Vials containing 0.1 ml of Muddy at 1 ×1011 PFU/ml were resuspended in 10 ml of Ringers Solution. One ml of this solution was used for each dose (1 ×109 PFU). Each batch of phage prepared was titered to check for correct concentration, subjected to an Endotoxin assay to look for any contaminating lipopolysaccharide, and sent to Accugen Laboratories for sterility testing.
Phage neutralization assays
Plasma or serum samples were incubated with phage by adding 10 µl of plasma or serum to 90 µl of phage buffer containing ~1×1010 PFU of phage Muddy. After incubating at room temperature for 2 or 24 hours, 10-fold serial dilutions were prepared and 3 µl of each dilution spotted onto top agar overlays containing M. smegmatis mc2155. Plates were incubated at 37 °C for 24 hours.
Elisa
Enzyme-linked immunosorbant assays (ELISAs) were performed as described previously14. In brief, EIA microplates (Corning CLS3590) were coated overnight with 100 µl of coating buffer (carbonate-bicarbonate with pH = 9.6, Sigma C3041) or phage Muddy diluted to 5 ×109 pfu ml−1 in coating buffer. After incubation, washing, and blocking, 100 µl of diluted patient serum was added and incubated at 4 °C for ~20 hours. Anti-Muddy IgG was probed by incubation with goat Anti-Human IgG Fc (HRP) pre-adsorbed (Abcam ab98624) diluted 1:10,000, and detected following addition of 100 µl of 3,3′,5,5′-Tetramethylbenzidine (TMB) substrate (Sigma T0440), an 8 min incubation, and addition of 2 N H2SO4 to stop the reaction. Absorbance at 570 nm (background) was subtracted from absorbance at 450 nm (signal) and the difference was plotted against the serum dilution and fit with a fixed-baseline logistic curve; half-maximum IgG titers were determined from the curves.
Statistics and reproducibility
The Origin 2021b (64-bit) SR2 9.8.5.212 software was used for ELISA data analysis. Fixed-baseline logistic functions were used to fit the data, as shown in Fig. 3E. Half-maximal serum dilution is a parameter of these fits, and was inverted to yield half-maximal titer (Fig. 3F). ELISA curves in Fig. 3E are averages of two technical replicates, with error bars showing one standard deviation from the mean. Half-maximal titers for the two replicates are shown as white datapoints in Fig. 3F, with bars indicating the average of two replicates and error bars of one standard deviation.
The screen for infection by M. smegmatis phages (Fig. 3A) was performed three times and Fig. 3A shows representative data. The screen using lytically propagated prophages (Fig. 3B) and the killing/survival assays (Fig. 3C, D) were performed once. Neutralization assays were performed twice and representative data are shown in Fig. 3G.
Clinical procedures
This research protocol was approved by the Massachusetts General Hospital/Brigham and Women’s Hospital institutional review board and the U.S. Food and Drug Administration via a single patient expanded access application. The patient provided informed consent, according to CARE guidelines and in compliance with the Declaration of Helsinki principles
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request. Source data are provided with this paper.
References
Wallace, R. J., Brown, B. A. & Onyi, G. O. Skin, soft tissue, and bone infections due to Mycobacterium chelonae chelonae: importance of prior corticosteroid therapy, frequency of disseminated infections, and resistance to oral antimicrobials other than clarithromycin. J. Infect. Dis. 166, 405–412 (1992).
Dousa, K. M. et al. Ibrutinib therapy and Mycobacterium chelonae skin and soft tissue infection. Open Forum Infect. Dis. 5, ofy168 (2018).
Wallace, R. J. et al. Spectrum of disease due to rapidly growing mycobacteria. Rev. Infect. Dis. 5, 657–679 (1983).
Uslan, D. Z., Kowalski, T. J., Wengenack, N. L., Virk, A. & Wilson, J. W. Skin and soft tissue infections due to rapidly growing mycobacteria: Comparison of clinical features, treatment, and susceptibility. Arch. Dermatol. 142, 1287–1292 (2006).
Kennedy, B. S. et al. Outbreak of Mycobacterium chelonae infection associated with tattoo ink. N. Engl. J. Med 367, 1020–1024 (2012).
Safranek, T. J. et al. Mycobacterium chelonae wound infections after plastic surgery employing contaminated gentian violet skin-marking solution. N. Engl. J. Med. 317, 197–201 (1987).
Inkster, T., Peters, C., Seagar, A. L., Holden, M. T. G. & Laurenson, I. F. Investigation of two cases of Mycobacterium chelonae infection in haemato-oncology patients using whole-genome sequencing and a potential link to the hospital water supply. J. Hosp. Infect. 114, 111–116 (2021).
Meyers, H. et al. An outbreak of Mycobacterium chelonae infection following liposuction. Clin. Infect. Dis. 34, 1500–1507 (2002).
Cooper, J. F., Lichtenstein, M. J., Graham, B. S. & Schaffner, W. Mycobacterium chelonae: a cause of nodular skin lesions with a proclivity for renal transplant recipients. Am. J. Med. 86, 173–177 (1989).
Brown-Elliott, B. A., Wallace, R. J., Blinkhorn, R., Crist, C. J. & Mann, L. B. Successful treatment of disseminated Mycobacterium chelonae infection with linezolid. Clin. Infect. Dis. 33, 1433–1434 (2001).
Wallace, R. J. et al. Genetic basis for clarithromycin resistance among isolates of Mycobacterium chelonae and Mycobacterium abscessus. Antimicrob. Agents Chemother. 40, 1676–1681 (1996).
Tebas, P., Sultan, F., Wallace, R. J. & Fraser, V. Rapid development of resistance to clarithromycin following monotherapy for disseminated mycobacterium chelonae infection in a heart transplant patient. Clin. Infect. Dis. 20, 443–444 (1995).
Dedrick, R. M. et al. Mycobacterium abscessus strain morphotype determines phage susceptibility, the repertoire of therapeutically useful phages, and phage resistance. MBio 12, e03431–20 (2021).
Dedrick, R. M. et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 25, 730–733 (2019).
Dedrick, R. M. et al. Potent antibody-mediated neutralization limits bacteriophage treatment of a pulmonary Mycobacterium abscessus infection. Nat. Med. 27, 1357–1361 (2021).
Shield, C. G. et al. Application of bacteriophages for mycobacterial infections, from diagnosis to treatment. Microorganisms 9, 2366 (2021).
Gordillo Altamirano, F. L. & Barr, J. J. Phage therapy in the postantibiotic era. Clin. Microbiol. Rev. 32, e00066–18 (2019).
Kakasis, A. & Panitsa, G. Bacteriophage therapy as an alternative treatment for human infections. A comprehensive review. Int. J. Antimicrob. Agents 53, 16–21 (2019).
Schooley, R. T. et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob. Agents Chemother. 61, e00954–17 (2017).
Law, N. et al. Successful adjunctive use of bacteriophage therapy for treatment of multidrug-resistant Pseudomonas aeruginosa infection in a cystic fibrosis patient. Infection 47, 665–668 (2019).
Labrie, S. J., Samson, J. E. & Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327 (2010).
Łusiak-Szelachowska, M. et al. Phage neutralization by sera of patients receiving phage therapy. Viral Immunol. 27, 295 (2014).
Hatfull, G. F., Dedrick, R. M. & Schooley, R. T. Phage therapy for antibiotic-resistant bacterial infections. Annu. Rev. Med. 73, 197–211 (2022).
Acknowledgements
We acknowledge Dr. Francisco M. Marty for inspiring the decision to pursue bacteriophage therapy for this patient as well as for his extensive efforts in the early clinical planning and regulatory process. We continue to mourn his death, may he rest in peace. We thank Deborah Jacobs-Sera and Lawrence A. Abad for phage preparation. This work was supported by grants GM131729 from the National Institutes of Health (GFH), GT12053 from the Howard Hughes Medical Institute (GFH), HATFUL19GO from the Cystic Fibrosis Foundation (GFH), and by a kind donation from The Fowler Fund for Phage Research (GFH).
Author information
Authors and Affiliations
Contributions
J.S.L., L.R.B., and D.A.S. designed the clinical protocol. CAB provided clinical guidance. R.M.D., K.G.F., M.M.C., B.E.S., and G.F.H. screened the bacteriophages, amplified and purified the final product, and tested patient sera for the development of antibodies. T.A.J. assisted with microbiologic management of the Mycobacterial isolate. R.M.D., K.G.F., M.M.C., B.E.S., and G.F.H. prepared Fig. 3. J.S.L. prepared Figs. 1 and 2 and wrote the primary manuscript and all other authors edited the final product. D.A.S. and G.F.H. contributed equally.
Corresponding author
Ethics declarations
Competing interests
C.A.B. receives contract and grant funding from Gilead. G.F.H. acts as consultant and grant recipient for Janssen pharmaceuticals. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Meritxell Garcia-Quintanilla, Ronen Hazan, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Little, J.S., Dedrick, R.M., Freeman, K.G. et al. Bacteriophage treatment of disseminated cutaneous Mycobacterium chelonae infection. Nat Commun 13, 2313 (2022). https://doi.org/10.1038/s41467-022-29689-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-022-29689-4
This article is cited by
-
Bacteriophage therapy for the treatment of Mycobacterium tuberculosis infections in humanized mice
Communications Biology (2024)
-
Therapeutically useful mycobacteriophages BPs and Muddy require trehalose polyphleates
Nature Microbiology (2023)
-
Phage Therapy for Nontuberculous Mycobacteria: Challenges and Opportunities
Pulmonary Therapy (2023)
-
Multiple drugs
Reactions Weekly (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.