Abstract
Macrophage-derived exosomes (Mφ-Exos) are involved in tumor progression, but its role in glioma is not fully understood. RBP-J is related to macrophage activation. In this study, we assess the role of exosomes derived from RBP-J-overexpressed macrophages (RBP-J OE Mφ-Exos) in glioma. The circular RNA (circRNA) profiles in RBP-J OE Mφ-Exos and THP-1-like macrophages (WT Mφ)-Exos were evaluated using circRNA microarray. Then the functions of Mφ-Exo-circRNA in glioma cells were assessed via CCK-8, EdU, Transwell invasion, and nude mouse assays. Besides, luciferase reporter assay, RNA immunoprecipitation, and Pearson’s correlation analysis were adopted to confirm interactions. We found that circRNA BTG (circBTG2) is upregulated in RBP-J OE Mφ-Exos compared to WT Mφ-Exos. RBP-J OE Mφ-Exos co-culture and circBTG2 overexpression inhibited proliferation and invasion of glioma cells, whereas circBTG2 knockdown promotes tumor growth in vivo. The effects of RBP-J OE Mφ-Exos on glioma cells can be reversed by the circBTG2 knockdown. In conclusions, Exo-circBTG2 secreted from RBP-J OE Mφ inhibits tumor progression through the circBTG2/miR-25-3p/PTEN pathway, and circBTG2 is probably a diagnostic biomarker and potential target for glioma therapy.
Similar content being viewed by others
Introduction
Gliomas are primary malignancy commonly seen in the nervous system [1, 2], which are featured with heterogeneous genetic molecular aberrations [3]. Glioblastoma multiforme (GBM) is the most aggressive type with repeated relapse. The median survival rate of GBM patients is only 14.6 months even after standard advanced surgery and chemoradiotherapy with temozolomide [4, 5]. Thus, top priority should be given to the probation of the possible molecular mechanisms by which gliomas progress, so as to improve the therapy of gliomas, especially GBM.
Increasing evidence unveils that the exosomes (Exos) mediate the interactions between macrophages and cancer cells [6,7,8]. M2 Macrophage-derived exosomes (Mφ-Exos) were found to boost cancer cells to migrate and invade [9], and tumor-associated Mφ-Exos facilitate gastric cancer cells to migrate via transfer of functional Apolipoprotein E [10]. Besides, Mφ-Exo-miR-501-3p contributes to progression of PDAC via the TGF-β signaling pathway mediated by TGFBR3 [11]. Downregulated lncRNA SBF2-AS1 in M2 Mφ-Exos raises miR-122-5p to restrict XIAP, thus curbing PC development [12]). M2 bone marrow-derived Mφ-Exos elevate miR-21 to accelerate immune escape of gliomas via modulating PEG3 [13].
Circular RNAs (circRNAs) have also been found in Exos [14, 15], and they are thought to modulate the expression of genes and miRNAs [16]. Exo-circRNAs can promote malignant phenotype of peripheral cells in cholangiocarcinoma [17]. Recent studies have unveiled the involvement of circRNAs in glioma progression by competitive sponging miRNAs [18, 19]. For instance, circ_0037655 is able to boost gliomas to progress via controlling miR-214/PI3K signal transduction [20]. However, whether Mφ-Exo-circRNAs can regulate the progression of gliomas is unclear.
The Notch pathway is involved in several core cellular processes, such as proliferation and tumor development [21, 22], and it is also believed to be responsible for the activation and differentiation of macrophages [23,24,25]. The recombination signal-binding protein-Jκ (RBP-J) is a transcriptional regulator that is often used as a marker for the activation of Notch signaling [26]. Notch intracellular domains are released by Notch ligands and translocate to the nucleus where they bind to RBP-J [27]. Loss of the Notch effector RBP-J promotes tumorigenesis [28]. Moreover, Notch-RBP-J signal transduction regulates the transcription factor IRF8 to facilitate inflammatory macrophage polarization [29].
In the current research, we probed the impacts of RBP-J OE Mφ-Exos (exosomes derived from RBP-J-overexpressed macrophages) on glioma cell proliferation and invasion and compared them with Exos from THP-1-like macrophages (WT Mφ-Exos) [30]. To further understand the regulatory mechanism of RBP-J OE Mφ-Exos in gliomas, we also determined the differentially regulated circRNAs when RBP-J was upregulated in Mφ-Exos. In addition, we identified the miRNA binding partners of the circRNA and their targets. This study aimed at identifying pathways that are uniquely expressed in glioma progression to understand the mechanisms of Mφ-Exo-circRNAs, and determining diagnostic biomarkers and potential therapeutic targets.
Results
RBP-J is lowly expressed in the macrophages from the glioma tissues and related to the prognosis of glioma patients
Glioma tissues (n = 40) and para-tumor tissues (n = 40) were obtained from 40 patients. The expression of RBP-J in the macrophages from the glioma tissue samples and adjacent normal tissue samples was detected using qRT-PCR. Results showed that RBP-J is lowly expressed in the macrophages from the glioma tissues (Fig. 1A). The correlation between RBP-J expression in the macrophages of glioma tissue samples and clinicopathological parameters of glioma patients is shown in Table 1. The median expression level of RBP-J in the macrophages of glioma tissues was taken as the cut-off value for high and low levels. We found that higher RBP-J expression was associated with lower glioma Grade (P < 0.05), but there was no association between the expression level of RBP-J and other clinicopathological parameters in glioma such as age and gender (P > 0.05). In addition, the survival rate of glioma patients with highly expressed RBP-J in the macrophages of glioma tissues was better than that of patients with lower RBP-J expression in the macrophages of glioma tissues (Fig. 1B).
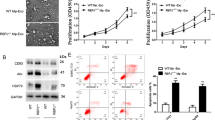
A RBP-J expression in the microphages from glioma tissues and paired normal tissue samples (n = 40) using qRT-PCR. B Kaplan–Meier analysis of overall survival of glioma patients stratified by RBP-J expression. C Cell proliferation in glioma cell lines U87 MG co-cultured with WT Mφ, RBP-J OE Mφ, spaced RBP-J OE Mφ, spaced RBP-J OE Mφ-EVs or RBP-J OE Mφ-supernatant was assessed by CCK-8. D Wound healing assay is performed to detect cell migration (bar = 100 μm). All experiments were performed three times. **P < 0.01, ***P < 0.001 for statistical differences, EVs Extracellular vesicles, ns no significance.
RBP-J OE Mφ inhibit proliferation and invasion of glioma cells through extracellular vesicles
To further investigate the effects of the RBP-J OE Mφ on the proliferation and migration of glioma cells, we cocultured U87 MG cells with Mφ for 5 days and measured cell proliferation and migration with CCK-8 and wound healing assays. As demonstrated in Fig. 1C, D, RBP-J OE Mφ significantly inhibited cell proliferation and migration of U87 MG cells compared with WT Mφ. We wonder whether Mφ play a role through direct contact or indirect contact. Thus, we co-culture Mφ and U87 cells in separate spaces, allowing only the medium to contact each other. Then we found that RBP-J OE co-culture with Mφ could also significantly inhibited cell proliferation and migration of U87 MG cells (Fig. 1C, D). This means that Mφ play a role through indirect contact. Nevertheless, whether it is through extracellular vesicles or soluble small molecules still needs to be further explored. We extracted extracellular vesicles and supernatant of Mφ respectively. As shown in Fig. 1C, D, we found that Mφ play a role through extracellular vesicles.
RBP-J OE Mφ-Exos inhibit proliferation and invasion of glioma cells
WT Mφ-Exos and RBP-J OE Mφ-Exos were isolated by ultracentrifugation and characterized by TEM and NTA (Fig. 2A). To further confirm the identity of the Exos, the expression levels of CD63 and TSG101 (Exo markers) were evaluated. Western blot assessment showed that the isolated Exos were enriched with CD63 and TSG101 (Fig. 2B). These data indicate the successful isolation of Exos from WT Mφ-Exos and RBP-J OE Mφ-Exos. Then, exosomes derived from WT Mφ and RBP-J OE Mφ cells were labeled with a green fluorescent marker, PKH67. After recipient cells (U87 MG cells) were incubated with labeled WT Mφ-Exos and RBP-J OE Mφ-Exos for 3 h, PKH67 was localized in the cytoplasm of recipient cells (Fig. 2C).
A Exosomes isolated from WT THP-1 derived macrophages (WT Mφ-Exo) and RBP-J-overexpressed macrophages (RBP-J OE Mφ-Exo) imaged by transmission electron microscopy (TEM) and their size distribution were measured using NTA. Scale bar = 50 nm. B Levels of exosome markers CD63 and TSG101 in WT or RBP-J OE Mφ-Exo were determined by Western blotting. C WT Mφ-Exo or RBP-J OE Mφ-Exo were labeled with PKH67; green represents PKH67, and blue represents nuclear DNA staining by DAPI. U87 MG cells were incubated with WT Mφ-Exo or RBP-J OE Mφ-Exo for 3 h. D, E Cell proliferation in glioma cell lines U87 MG and U373 MG treated with WT Mφ-Exo or RBP-J OE Mφ-Exo was assessed by CCK-8 (D) and EdU assay (E). F Transwell invasion assay is performed to indicate cell invasion (bar = 100 μm). All experiments were performed three times. *P < 0.05 and **P < 0.01 for statistical differences.
To further investigate the effects of these two groups of Exos on the proliferation of glioma cells, we cocultured U87 MG or U373 MG cells with Exos for 5 days and measured cell proliferation through CCK-8 and EdU experiments. As demonstrated in Fig. 2D, E, the presence of RBP-J OE Mφ-Exos significantly curbed U87 MG and U373 MG cells to proliferate when compared with WT Mφ-Exos. In addition, Transwell invasion assays indicated that RBP-J OE Mφ-Exos were able to inhibit invasion of U87 MG and U373 MG cells (Fig. 2F). Overall, these results confirm that the overexpression of RBP-J in Exos can suppress glioma cells to proliferate and invade.
Expression profiles of circRNAs in RBP-J OE Mφ-Exos
We wonder whether RBP-J OE Mφ-Exos could influence the expression of RBP-J in glioma cells. We performed qRT-PCR to detect the expression of RBP-J in the WT Mφ-Exos treated U87 MG cells, and RBP-J OE Mφ-Exos treated U87 MG cells. Results showed that there is no significant difference of RBP-J level between WT Mφ-Exos treated U87 MG cells and RBP-J OE Mφ-Exos treated U87 MG cells (Fig. 3A). This result means that RBP-J OE Mφ-Exos can’t influence the expression of RBP-J in U87 MG cells. Thus, we wonder whether there are different expressions of circRNAs between WT Mφ-Exos and RBP-J OE Mφ-Exos. The circRNA profiles in RBP-J OE Mφ-Exos and WT Mφ-Exos were evaluated using a circRNA microarray technique. 39 circRNAs were differentially expressed (P < 0.05 and log2FC > 2.0 or < −2.0) in RBP-J OE Mφ-Exos and the controls (Fig. 3B). Among them, 25 circRNAs dramatically rose up and 14 ones evidently declined. circBTG2 with the most obvious rising trend was selected and validated to be present in RBP-J OE Mφ-Exos and WT Mφ-Exos by qRT-PCR (Fig. 3C). In the meantime, it was unveiled that circBTG2 was expressed in the RBP-J OE Mφ at a notably higher level relative to that in the WT Mφ cells (Fig. 3D). Compared with those in the producer cells, the levels of circBTG2 are enriched by approximately 4 folds in the RBP-J OE Mφ-Exos and WT Mφ-Exos (Fig. 3E). This result means that circBTG2 is enriched in exosomes. According to the circBase (http://www.circbase.org), circBTG2 (chr1:203274663-203278729) was derived from BTG2.
A The relative expressions of RBP-J in WT Mφ-Exos treated U87 MG cells and RBP-J OE Mφ-Exos treated U87 MG cells were detected by qRT-PCR. B Cluster heatmap showing 39 aberrantly expressed circRNAs, including 25 upregulated and 14 downregulated circRNAs in exosomes derived from RBP-J-overexpressed macrophages compared to the controls. The red color represents high expression, whereas the green color represents low expression. C, D The relative expression of circRNA BTG2 in Mφ-Exo (C) and Mφ cells (D) was validated by qRT-PCR. E The fold change of circRNA BTG2 expression between the exosomes and their corresponding producer cells. F, G The qRT-PCR assay indicated the difference in the circRNA BTG2 expression in RBP-J-overexpressed Mφ cells transfected with or without si-circRNA (F), as well as in exosomes from those cells (G). **P < 0.01 and ***P < 0.001 for statistical differences, ns: no significance.
Mφ-Exo-circBTG2 inhibits glioma cells to proliferate and invade
Since the circRNA BTG2 level was the highest in RBP-J OE Mφ-Exos, to remove its expression from Exos, the siRNA of circBTG2 was transfected into Mφ cells for 48 h, after which Exos were collected (Fig. 3F, G). Next, glioma cell proliferation and invasion were investigated by coculturing cells with Mφ-Exos. The inhibitory effects of RBP-J OE Mφ-Exos on the proliferation and invasion of glioma cells (U87 MG and U373 MG) were eliminated when circBTG2 was knocked down in Mφ (Fig. 4A–C). This would be expected if there was an association between the expression of RBP-J and circBTG2.
To remove circRNA BTG2 from exosomes, siRNA of circRNA BTG2 was transfected into THP-1 cells and Mφ-Exo were collected at 48 h post-transfection (RBP-J OE Mφ-Exo-si-circRNA). Glioma cell lines U87 MG and U373 MG were cocultured with WT Mφ-Exo, RBP-J OE Mφ-Exo or (RBP-J OE + si-circRNA) Mφ-Exo. A, B Cell proliferation in glioma cell lines U87 MG and U373 MG was assessed by CCK-8 assay (A) and EdU assay (B). C Cell invasion in glioma cell lines U87 MG and U373 MG was assessed by Transwell assay (bar = 100 μm). All experiments were performed three times. *#P < 0.05 and ***###P < 0.001 as indicated. * vs. WT Mφ-Exo, # vs. RBP-J OE Mφ-Exo.
To continuously figure out the biological role of circBTG2 in glioma cells, U87 MG and U373 MG cells underwent transfection with a circBTG2 overexpression vector (Supplementary Fig. S1A). The results unveiled that circBTG2 overexpression significantly inhibited glioma cells to proliferate and invade (Supplementary Fig. S1B–D). As with the overexpression of RBP-J, the overexpression of circBTG2 inhibited proliferation and invasion of glioma cells.
circBTG2 acts as a sponge for miR-25-3p
For discovering more about the specific regulation of circBTG2, we performed bioinformatics prediction (starBase). Bioinformatics analysis predicted that circBTG2 and miR-25-3p possessed complementary binding sites (Fig. 5A). Then we carried out a dual-luciferase experiment in 293 T cells to confirm this interaction by mutating the predicted binding site in circBTG2. It was unveiled that the luciferase activity was reduced only in presence of WT circBTG2 and miR-25-3p mimics in 293 T cells (Fig. 5B), which was further validated using an Ago2 RIP assay. Ago2 significantly enriched RNA levels of both circBTG2 and miR-25-3p (Fig. 5C). Besides, levels of circBTG2 and miR-25-3p were also analyzed in glioma and matched para-carcinoma tissues, and the results further substantiated that glioma tissues exhibited a lower circBTG2 level (Fig. 5D) and a higher miR-25-3p level (Fig. 5E). Besides, Pearson’s analysis confirmed a negative interrelation between circBTG2 and miR-25-3p in glioma and matched para-carcinoma tissues (Fig. 5F). This indicates that circBTG2 may compete to bind to miR-25-3p as a sponge and prevent it from regulating other pathways.
A Putative complementary sites within miR-25-3p and circRNA BTG2 were predicted by bioinformatics analysis. B Dual-luciferase reporter assays demonstrate that miR-25-3p is a direct target of circRNA BTG2 in 293 T cells. C The Ago2 RIP showed that Ago2 significantly enriched circRNA BTG2 and miR-25-3p. D The expression level of circRNA BTG2 in 40 glioma tissues and matched para-carcinoma normal tissues was determined by qRT-PCR. E The expression level of miR-25-3p in the above tissues was determined by qRT-PCR. F The expression levels of miR-25-3p are negatively correlated with circRNA BTG2 in glioma tissues. **P < 0.01 and ***P < 0.001 for statistical differences.
circBTG2 represses glioma cells to proliferate and invade via the miR-25-3p/PTEN pathway
We next probed the potential binding sites of miR-25-3p. Target prediction and assessment were implemented using starBase (http://starbase.sysu.edu.cn) and miRDB (http://mirdb.org), which identified that miR-25-3p probably interacts with PTEN, a tumor suppressor gene implicated in several cancers [31, 32]. Later, we mutated two potential miR-25-3p target sites in PTEN (Fig. 6A) and performed a luciferase reporter experiment., which ascertained that miR-25-3p overexpression in HEK293T cells dramatically weakened the luciferase activity of PTEN at both target sites (Fig. 6B). Thereafter, we examined the transfection efficiency of miR-25-3p mimics and inhibitor (Fig. 6C). The mRNA and protein levels of PTEN dropped down in U87 MG and U373 MG cells transfected with miR-25-3p mimics but rose up in glioma cells undergoing miR-25-3p inhibitor transfection (Fig. 6D, E). Overexpression of circBTG2 upregulated PTEN whereas miR-25-3p mimics transfection reversed it in U87 MG cells (Fig. 6F). Relative to the matched para-carcinoma tissues, PTEN was expressed at a lower level in glioma tissues (Fig. 6G). In glioma tissues, PTEN was negatively correlated with miR-25-3p, but positively correlated with circBTG2 expression (Fig. 6H, I). Further, circBTG2 overexpression inhibited cells to proliferate and invade whereas miR-25-3p mimics transfection reversed it in U87 MG and U373 MG cells (Supplementary Fig. S2A–C). It can be assumed that circBTG2 inhibits proliferation and invasion in glioma cells by sponging miR-25-3p and upregulating PTEN expression.
A Bioinformatics analysis revealed the predicted binding sites between PTEN and miR-25-3p. B Luciferase reporter assay demonstrated miR-25-3p mimics significantly decreased the luciferase activity of PTEN-WT in HEK293T cells. C The transfection efficiency of miR-25-3p mimics and inhibitor in U87 MG and U373 MG cells. D, E The mRNA (D) and protein (E) level of PTEN was detected through qRT-PCR and western blotting after transfection with miR-25-3p mimics and inhibitor in U87 MG and U373 MG cells. F circRNA BTG2 overexpression upregulated PTEN, this effect can be reversed by co-transfection with miR-25-3p mimics in U87 MG cells. G The expression levels of PTEN in 40 glioma tissues and matched para-carcinoma normal tissues was determined by qRT-PCR. H Expression levels of PTEN negatively correlated with miR-25-3p in glioma tissues. I Expression levels of PTEN positively correlated with circRNA BTG2 in glioma tissues. **P < 0.01 and ***P < 0.001 for statistical differences.
RBP-J OE Mφ-Exo inhibits tumor growth through the circBTG2/miR-25-3p/PTEN pathway in vivo
For proving the effect of Mφ-Exo-circBTG2 on the modulation of glioma growth in vivo, U87 MG cells undergoing transfection with sh-circRNA or sh-NC or coculture with WT Mφ-Exos, RBP-J OE Mφ-Exos or (RBP-J OE + sh-circRNA) Mφ-Exo were subcutaneously injected into nude mice. Tumors cultured with RBP-J OE Mφ-Exos were significantly smaller, whereas those undergoing sh-circRNA transfection were significantly larger. The greatest differences in the tumor volume and weight were observed in the tumors between RBP-J OE Mφ-Exo group and sh-circRNA group (Fig. 7A–C). What’s more, the inhibitory effects of RBP-J OE Mφ-Exos on the tumor growth in vivo were eliminated when circRNA BTG2 was knocked down in Mφ (Fig. 7A–C). The relative expression of circBTG2 was the highest in the RBP-J OE Mφ-Exo group and the lowest in sh-circRNA group (Fig. 7D). While the relative expression of miR-25-3p was the lowest in the RBP-J OE Mφ-Exo group and the highest in sh-circRNA group (Fig. 7E). Meanwhile, protein levels of PTEN were the highest in RBP-J OE Mφ-Exo group and lowest in sh-circRNA group (Fig. 7F). These results signify that RBP-J OE Mφ-Exos might inhibit tumor growth through a circBTG2/miR-25-3p/PTEN pathway in xenograft tumor models.
A Representative images of xenograft tumors (three mice per group) in nude mice. B Tumor volume is monitored every 7 days for 35 days. C The weights of xenograft tumors are summarized. D qRT-PCR verification of the expression of circRNA BTG2 in tumors (n = 3). E qRT-PCR verification of the expression of miR-25-3p in tumors (n = 3). F The protein expression of PTEN was detected by western blotting in tumors (n = 3). *P < 0.05, **P < 0.01, and ***P < 0.001 for statistical differences.
Discussion
Macrophages are abundant in the glioma tumoral environment and associated with chronic inflammation [33, 34]. Moreover, the macrophage environment is heterogenous with the progression of tumors dependent on alternatively polarized M2 macrophages and tumorigenic immune responses dependent on M1-polarized macrophages [35, 36]. Therefore, improving the understanding of macrophage regulation in the tumoral environment is important in developing effective therapies for gliomas. Notch-RBP-J signaling is believed to regulate TLR-induced inflammatory macrophage polarization by the indirect regulation of M1-specific genes [29].
In this study, we examined whether RBP-J overexpression in macrophages would influence glioma cells. We found that RBP-J OE Mφ-Exos could curb glioma cells to proliferate and invade. Furthermore, we probed their interrelations by investigating the differentially regulated circRNAs in Mφ-Exos with upregulated RBP-J. Using the circRNA microarray technique, we discovered that 39 Exo-circRNAs were differentially regulated in WT Mφ-Exos with RBP-J overexpression, of which 25 were upregulated and 14 were downregulated. Later, we selected the Exos with the highest circBTG2 expression for further analysis. Then we unveiled that the inhibitory effects of RBP-J OE Mφ-Exos on the proliferation and invasion of glioma cells (U87 MG and U373 MG) were eliminated when circBTG2 was knocked down in Exos. Meanwhile, circBTG2 overexpression dramatically repressed glioma cells to proliferate and invade. These associations required further investigation, so we searched for miRNAs that may interact with circBTG2.
The public database (starBase) predicted that circBTG2 may interact with miR-25-3p, which was validated via luciferase or RIP assays. Our studies proved that the overexpression of circBTG2 could reduce miR-25-3p level. Then, a negative correlation between miR-25-3p and circBTG2 in glioma and matched para-carcinoma tissues was confirmed by Pearson’s analysis. Thus, we deduced that circBTG2 may repress miR-25-3p to prevent it from interacting in other pathways. StarBase revealed that miR-25-3p interacted with PTEN, a well-known tumor suppressor gene [31, 32]. Mutation or deletion of PTEN via complete loss of its locus on chromosome 10q is found in a multifold of GBMs [37, 38] and correlated with poor prognosis in diverse glioma subtypes [39, 40]. Furthermore, PTEN loss dramatically enhances gliomagenesis in quantities of murine model systems [41,42,43]. In the current research, we found that circBTG2 overexpression upregulated PTEN and inhibited cells to proliferate and invade whereas miR-25-3p mimics transfection reversed them in glioma cells. What’s more, PTEN expression displayed a negative interrelation with miR-25-3p level and a positive correlation with circBTG2 level in glioma tissues. In vivo assays further verified that RBP-J OE Mφ-Exos might inhibit tumor growth through a circBTG2/miR-25-3p/PTEN pathway in xenograft tumor models.
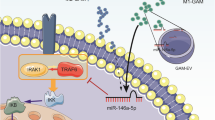
To conclude, circBRG2 suppress glioma cells to proliferate and invade via the miR-25-3p/PTEN pathway. Moreover, RBP-J OE Mφ-Exo inhibits tumor growth by stimulating the circBRG2/miR-25-3p/PTEN pathway in vitro and in vivo (Fig. 8). The above results indicate that RBP-J OE Mφ-Exos probably play a potential regulation role in the glioma progression and circBTG2 could be a biomarker for glioma diagnosis and potential target for glioma therapy.
Materials and methods
Cell culture and clinical specimens
Human monocytic cell line THP-1 and glioma cell lines (U87 MG and U373 MG) were purchased from ATCC and maintained according to ATCC guidelines. THP-1 cells were cultured in RPMI-1640 medium provided by Gibco (Shanghai, China), and glioma cells were cultured in Dulbecco’s Modified Eagle medium (DMEM, Gibco, China) with 10% heat-inactivated fetal bovine serum (FBS) from Thermo Fisher Scientific (Shanghai, China), 100 U/mL penicillin, and 100 µg/mL streptomycin from HyClone Laboratories (Beijing, China) at 37 °C in a moist incubator with 5% CO2 and used in the exponential growth phase.
Forty paired glioma tissues and para-tumor tissues were obtained from patients receiving surgery at The First people’s Hospital of Yancheng between 2015 and 2018. They were diagnosed by histopathology and received no treatment prior to the operation. Besides, all participants signed informed consent in written form before the research. This research gained the approval of the Ethics Committees of The First people’s Hospital of Yancheng and was conducted as per the Helsinki Declaration.
Macrophage extraction
The tissue single cell suspension (2*108/mL) was obtained and mixed into the separation solution (Sangon Biotech, Shanghai, China). The mixture was centrifuged for 20 min (1500 r/min) and the milky white macrophage layer was then collected. The macrophage layer was added into a test tube containing 5 mL of cell washing solution. After mixing, the macrophage layer was centrifuged (1800 r/min) and washed twice.
Isolation of Exos derived from THP-1 Mφ cells with or without the overexpression of RBP-J
To obtain WT Mφ and RBP-J OE Mφ, THP-1 cells underwent transfection with the pCMV6 empty vector or pCMV6 overexpressing RBP-J (OriGene, Rockville, MD, USA) and seeded at 1 × 106 cells/well in a six-well culture plate. Cellular debris were discarded from the collected culture medium that was centrifuged at 3000 × g for 15 min. Then Exoquick exosome precipitation solution (System Biosciences, CA, U.S.A.) was utilized for exosome separation [44,45,46].
Nanoparticle tracking analysis (NTA)
We measured the exosome particle size and concentration using NTA with ZetaView PMX 110 (Particle Metrix, Meerbusch, Germany) and corresponding software ZetaView 8.04.02. Isolated exosome samples were appropriately diluted using 1X PBS buffer (Biological Industries, Israel) to measure the particle size and concentration. NTA measurement was recorded and analyzed at 11 positions. The ZetaView system was calibrated using 110 nm polystyrene particles. The temperature was maintained at around 23 °C and 37 °C.
Transmission electron microscopy assay
Exos for transmission electron microscopy (TEM) were prepared as mentioned above.32 Briefly, Exos were first fixed in 2.5% glutaraldehyde (pH 7.2) at 4 °C, then washed in PBS, embedded in 10% gelatin and fixed in 1% osmium tetroxide for 60 min at indoor temperature. Next, the embedded Exos were cut into 1 mm-thick blocks and dehydrated with gradient alcohol. The alcohol was then replaced with gradient mixture of Quetol-812 epoxy resin and propylene oxide. Afterward, samples were embedded in Quetol-812 epoxy resin, polymerized at a temperature gradient, and cut into ultrathin sections using a Leica UC6 ultramicrotome. Finally, subsequent to dying by uranyl acetate and lead citrate, a transmission electron microscope was utilized for section observation.
Exosome labeling
Exosomes from 1.5 × 106 cells were suspended in 100 μl of PBS with 1 ml of mixed PKH67 (Sigma, in Diluent C). After 4 min of incubation at room temperature, 2 ml of 0.5% bovine serum albumin (BSA) was added to terminate exosome labeling, and dyed exosomes were isolated using Exoquick exosome precipitation solution. Exosomes were suspended in 9.6 ml of basal medium, and 250 μl was added to the sub-confluent layer of U87 MG cells. After incubation for 3 h at 37 °C, cells were washed and fixed at room temperature. To stain the nuclei, 4′,6-diamidino-2-phenylindole (DAPI, Sigma) was added for 10 min, and the stained cells were observed with a fluorescence microscope (Zeiss, LSM700B, Germany).
Microarray analysis
The isolation and quantification of the total RNAs were independently implemented using Trizol reagent provided by Life Technologies (Shanghai, China) and NanoDrop ND-1000. To enrich circRNAs and remove linear RNAs, RNase R provided by Epicenter Biotechnologies (Beijing, China) was utilized for RNA digestion. The enriched circRNAs were then amplified and labeled fluorescently using a Super RNA Labeling kit from Arraystar (Shanghai, China) as per the guideline of the manufacturer. In the meantime, we hybridize the labeled cRNAs onto an Arraystar Human circRNA Array V2 (8 × 15 K). Subsequent to rinsing and scanning of slides using an Agilent Scanner G2505C, the obtained images were assessed via Agilent Feature Extraction software (version11.0.1.1). Later, the limma package in R was utilized for quantile normalization and data assessment. Ultimately, fold change filtering and hierarchical clustering were used to determine circRNAs with differential expressions and their expression patterns.
RNA extraction and quantitative real-time PCR (qRT-PCR)
The reverse transcription of mRNAs and circRNAs into cDNAs was implemented using a reverse transcription kit from Takara (Beijing, China), which were synthesized using the miRNA 1st-strand cDNA synthesis kit (Sangon Biotech, China). Next, cDNAs were subjected to RT-PCR on a Quantstudio™ DX system (Applied Biosystems, Singapore) under the following conditions: denaturation at 95 °C for 30 s and (denaturation at 95 °C for 5 s, at 60 °C for 10 s and at 72 °C for 30 s) × 40 cycles. Afterward, we utilized 2-ΔΔCT to quantify mRNAs and circRNAs by normalizing to GAPDH [47] and to determine the relative expression subsequent to the normalization of miRNA expression to small nuclear U6. Each experiment was separately performed in triplicate. All PCR primers were listed in Table 2.
Cell transfection
CircRNA BTG anti-proliferation factor 2 (BTG2) (circBTG2) overexpression plasmid (p-circRNA) and its mimic pcDNA3.1, small interfering RNAs (siRNAs) targeting circRNAs and nonspecific negative control oligos (si-NC), miR-25-3p mimics, inhibitor and the negative control (NC), and the lentivirus targeting circBTG2 were bought from GeneChem (Shanghai, China). Detailed sequences were depicted in Table 2. U87 MG and U373 MG cell lines underwent inoculation in six-well plates at 24 h prior to transfection with pcDNA3.1, p-circRNA, si-NC, si-circRNA, and miR-25-3p mimics or inhibitor under 50–60% cell confluence using Lipofectamine 3000 (Invitrogen) as per the guideline of the manufacturer. Later, the effects of knockdown or overexpression were examined by qRT-PCR using the RNAs that were extracted after 48-h transfection. For Exo treatment, glioma cells were cultured in medium containing 5 μg/ml Exos from WT Mφ, RBP-J OE Mφ or (si-circRNA + RBP-J OE) Mφ.
Cell proliferation assays
Approximately 1.0 × 103 transfected U87 MG and U373 MG cells were cultured in 96-well plates, and then underwent 1 h incubation with CCK-8 reagent (Beyotime, Shanghai, China). The absorbance at 450 nm was recorded using an Infinite M200 multimode microplate reader (Tecan, Shanghai, China).
After approximately 48 h transfection, the 5-ethynyl-2’-deoxyuridine (EdU) assay kit provided by Ribo (Guangzhou, China) was utilized to examine the proliferation of U87 MG and U373 MG cells. Specifically, cells were grown in culture medium containing EdU (Invitrogen) solution (1:1000). At the proliferative stage, the cells were labeled with EdU for 2 h, followed by rinsing with PBS (0.5 g/mL) thrice. Subsequently, the cells were stained by 4′,6-diamidino-2-phenylindole (DAPI) from Invitrogen for 10 min at indoor temperature in the dark and underwent PBS rinsing more than twice. Ultimately, assessment of the stained cells was implemented via the FACSCalibur DxP flow cytometer (BD Biosciences, Shanghai, China).
Cell invasion assays
For invasion assays, the lower chambers were precoated with 100 μL of Matrigel (BD Bioscience, San Jose, CA, USA) for 30 min before the addition of medium to the chambers. The glioma cells (2 × 105 cells/mL) were resuspended in DMEM medium. The upper chamber contained 100 μL of cell suspension medium, and 600 μL of complete medium was added to the bottom chamber. After incubating at 37 °C with 5% CO2 for 24 h, cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet solution. The cells that passed through the filter were photographed and counted by inverted fluorescence microscopy (Leica Microsystems GmbH, Wetzlar, Germany) in four randomly selected fields.
Luciferase reporter assay
Sequences of WT or MUT circBTG2 or the full length of the 3′-UTR of PTEN with WT or MUT putative binding sites were interposed into the pmir-GLO vector from Promega Corp. (Beijing, China). 293 T cells seeded into 24-well plates underwent co-transfection with 50 nM miR-25-3p mimics or a NC and 80 ng WT or MUT plasmids using Lipofectamine 2000 (Invitrogen)and the 80 ng of plasmids were later added with 5 ng of pRL-SV40. Lastly, luciferase intensity was determined using the Dual-Luciferase Reporter Assay Kit from Promega (Beijing, China) and a microplate reader.
RNA binding protein immunoprecipitation (RIP) assay
We carried out the RIP assay using a Magna RIP Kit from Millipore (Hongkong, China) as per the guideline of the manufacturer. Specifically, cells (2 × 107) were lysed with the lysis buffer provided in the kit and the lysate was separately put into two tubes [one with anti-Argonaute2 (AGO2) antibody and the other with a nonspecific anti-IgG antibody (Millipore)]. The cell lysates were incubated nightlong at 4 °C, and then incubated with magnetic beads for a further hour. Proteinase K was then added for sample incubation at 55 °C for another hour. In the end, RNA extraction reagent (Solarbio, Beijing, China) was used to obtain the RNAs, and specific genes were detected and measured using qRT-PCR.
Western blot analysis
Cell lysis was performed in RIPA buffer (Beyotime, Nantong, China) containing protease and phosphatase inhibitors (Beyotime). A BCA Protein Assay kit (Beyotime) was utilized to identify protein concentration, and the samples (40 µg proteins per lane) underwent SDS-PAGE with 10% gel for separation. Next, proteins were electrotransferred onto a PVDF membrane (Beyotime) that was sealed by 5% BSA (Beyotime) for 1 h at indoor temperature. Later, we incubated the membrane with primary antibodies against TSG101 (1:1000, ab125011, Abcam, Shanghai, China), CD63 (1:1000, ab217345, Abcam, Shanghai, China), PTEN (1:1000, ab267787, Abcam, Shanghai, China) and GAPDH (1:1000, ab8245, Abcam, Shanghai, China) at 4 °C nightlong. The membranes were then probed with HRP-labeled secondary antibodies (Beyotime, Nantong, China) at indoor temperature for 1 h and signals were detected by chemiluminescence.
Xenograft nude mouse model
6–8-week-old adult male BALB/C nude mice (n = 3/group) were commercially provided by Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China) and reserved in a SPF environment with a LD (12: 12) cycle. All animal studies obtained the approval of the Institutional Animal Care and Use Committee of The First people’s Hospital of Yancheng and implemented in line with institutional and national guidelines. U87 MG cells undergoing stable sh-NC or sh-circRNA transfection, or WT Mφ-Exo, RBP-J OE Mφ-Exo or RBP-J OE Mφ-Exo-sh-circRNA (5 μg/ml) treatment were hypodermically injected into the nude mice (1 × 106 cells per mouse) on the right upper back. Later, we utilized a caliper to determine the growth of tumor every 7 days for 35 days, and calculate its volume based on the formula: volume = (length × width2) /2. Five weeks later, we intraperitoneally injected overdose pentobarbital (>120 mg/kg body weight) to kill all the mice so that they were unable to spontaneously breath. Afterward, the xenograft tumor tissues were sampled for subsequent analyses.
Statistical assessment
GraphPad Prism 6.0 software provided by GraphPad Inc. (San Diego, CA, USA) was utilized to statistically evaluate data. Experimental results were presented as mean ± standard deviation (SD). The statistically significant differences between tumor tissues and para-tumor tissues were determined using paired Student’s t-test. Besides, the statistically significant differences between other two groups were detected using Mann–Whitney U-test or unpaired Student’s t-test in light of conditions. Furthermore, the comparisons among different groups (multigroup comparisons) were implemented by one-way ANOVA and the post hoc Bonferroni test. Lastly, Pearson’s correlation coefficient was determined to test associations among circBTG2, miR-25-3p, and PTEN. Log-rank test and Kaplan–Meier method were used to assess survival rates. Data concerning the association of RBP-J expression with clinicopathological features of glioma were analyzed by Fisher’s exact test. P < 0.05 signified statistically significant differences.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Grasso CS, Tang Y, Truffaux N, Berlow NE, Liu L, Debily MA, et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med. 2015;21:555–9.
Lin GL, Nagaraja S, Filbin MG, Suva ML, Vogel H, Monje M. Non-inflammatory tumor microenvironment of diffuse intrinsic pontine glioma. Acta Neuropathol Commun. 2018;6:51.
Koul D, Wang S, Wu S, Saito N, Zheng S, Gao F, et al. Preclinical therapeutic efficacy of a novel blood-brain barrier-penetrant dual PI3K/mTOR inhibitor with preferential response in PI3K/PTEN mutant glioma. Oncotarget. 2017;8:21741–53.
Lu F, Chen Y, Zhao C, Wang H, He D, Xu L, et al. Olig2-dependent reciprocal shift in PDGF and EGF receptor signaling regulates tumor phenotype and mitotic growth in malignant glioma. Cancer Cell. 2016;29:669–83.
Akay M, Hite J, Avci NG, Fan Y, Akay Y, Lu G, et al. Drug screening of human GBM spheroids in brain cancer chip. Sci Rep. 2018;8:15423.
Cai J, Qiao B, Gao N, Lin N, He W. Oral squamous cell carcinoma-derived exosomes promote M2 subtype macrophage polarization mediated by exosome-enclosed miR-29a-3p. Am J Physiol Cell Physiol. 2019;316:C731–40.
Ham S, Lima LG, Chai EPZ, Muller A, Lobb RJ, Krumeich S, et al. Breast cancer-derived exosomes alter macrophage polarization via gp130/STAT3 signaling. Front Immunol. 2018;9:871.
Biswas S, Mandal G, Roy Chowdhury S, Purohit S, Payne KK, Anadon C, et al. Exosomes produced by mesenchymal stem cells drive differentiation of myeloid cells into immunosuppressive M2-polarized macrophages in breast cancer. J Immunol. 2019;203:3447–60.
Lan J, Sun L, Xu F, Liu L, Hu F, Song D, et al. M2 macrophage-derived exosomes promote cell migration and invasion in colon cancer. Cancer Res. 2019;79:146–58.
Zheng P, Luo Q, Wang W, Li J, Wang T, Wang P, et al. Tumor-associated macrophages-derived exosomes promote the migration of gastric cancer cells by transfer of functional Apolipoprotein E. Cell Death Dis. 2018;9:434.
Yin Z, Ma T, Huang B, Lin L, Zhou Y, Yan J, et al. Macrophage-derived exosomal microRNA-501-3p promotes progression of pancreatic ductal adenocarcinoma through the TGFBR3-mediated TGF-beta signaling pathway. J Exp Clin Cancer Res. 2019;38:310.
Yin Z, Zhou Y, Ma T, Chen S, Shi N, Zou Y, et al. Down-regulated lncRNA SBF2-AS1 in M2 macrophage-derived exosomes elevates miR-122-5p to restrict XIAP, thereby limiting pancreatic cancer development. J Cell Mol Med. 2020;24:5028–38.
Yang F, Wang T, Du P, Fan H, Dong X, Guo H. M2 bone marrow-derived macrophage-derived exosomes shuffle microRNA-21 to accelerate immune escape of glioma by modulating PEG3. Cancer Cell Int. 2020;20:93.
Zhu Z, Li Y, Liu W, He J, Zhang L, Li H, et al. Comprehensive circRNA expression profile and construction of circRNA-associated ceRNA network in fur skin. Exp Dermatol. 2018;27:251–7.
Su Y, Lv X, Yin W, Zhou L, Hu Y, Zhou A, et al. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging (Albany NY). 2019;11:8182–203.
Su Y, Xu C, Liu Y, Hu Y, Wu H. Circular RNA hsa_circ_0001649 inhibits hepatocellular carcinoma progression via multiple miRNAs sponge. Aging (Albany NY). 2019;11:3362–75.
Wang S, Hu Y, Lv X, Li B, Gu D, Li Y, et al. Circ-0000284 arouses malignant phenotype of cholangiocarcinoma cells and regulates the biological functions of peripheral cells through cellular communication. Clin Sci (Lond). 2019;133:1935–53.
Guan Y, Cao Z, Du J, Liu T, Wang T. Circular RNA circPITX1 knockdown inhibits glycolysis to enhance radiosensitivity of glioma cells by miR-329-3p/NEK2 axis. Cancer Cell Int. 2020;20:80.
Jin T, Liu M, Liu Y, Li Y, Xu Z, He H, et al. Lcn2-derived circular RNA (hsa_circ_0088732) inhibits cell apoptosis and promotes EMT in glioma via the miR-661/RAB3D axis. Front Oncol. 2020;10:170.
Qiao J, Liu M, Tian Q, Liu X. Microarray analysis of circRNAs expression profile in gliomas reveals that circ_0037655 could promote glioma progression by regulating miR-214/PI3K signaling. Life Sci. 2020;245:117363.
Ye YC, Zhao JL, Lu YT, Gao CC, Yang Y, Liang SQ, et al. NOTCH signaling via WNT regulates the proliferation of alternative, CCR2-independent tumor-associated macrophages in hepatocellular carcinoma. Cancer Res. 2019;79:4160–72.
Zhao JL, Huang F, He F, Gao CC, Liang SQ, Ma PF, et al. Forced activation of notch in macrophages represses tumor growth by upregulating miR-125a and disabling tumor-associated macrophages. Cancer Res. 2016;76:1403–15.
Pagie S, Gerard N, Charreau B. Notch signaling triggered via the ligand DLL4 impedes M2 macrophage differentiation and promotes their apoptosis. Cell Commun Signal. 2018;16:4.
Xu J, Chi F, Tsukamoto H. Notch signaling and M1 macrophage activation in obesity-alcohol synergism. Clin Res Hepatol Gastroenterol. 2015;39:S24–8.
Huang F, Zhao JL, Wang L, Gao CC, Liang SQ, An DJ, et al. miR-148a-3p mediates notch signaling to promote the differentiation and M1 activation of macrophages. Front Immunol. 2017;8:1327.
Tanigaki K, Kuroda K, Han H, Honjo T. Regulation of B cell development by Notch/RBP-J signaling. Semin Immunol. 2003;15:113–9.
Hsieh JJ, Henkel T, Salmon P, Robey E, Peterson MG, Hayward SD. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–9.
Kulic I, Robertson G, Chang L, Baker JH, Lockwood WW, Mok W, et al. Loss of the Notch effector RBPJ promotes tumorigenesis. J Exp Med. 2015;212:37–52.
Xu H, Zhu J, Smith S, Foldi J, Zhao B, Chung AY, et al. Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nat Immunol. 2012;13:642–50.
Auwerx J. The human leukemia cell line, THP-1: a multifacetted model for the study of monocyte-macrophage differentiation. Experientia. 1991;47:22–31.
Stewart DJ, Nunez MI, Jelinek J, Hong D, Gupta S, Aldaz M, et al. Impact of decitabine on immunohistochemistry expression of the putative tumor suppressor genes FHIT, WWOX, FUS1 and PTEN in clinical tumor samples. Clin Epigenetics. 2014;6:13.
Frew IJ, Thoma CR, Georgiev S, Minola A, Hitz M, Montani M, et al. pVHL and PTEN tumour suppressor proteins cooperatively suppress kidney cyst formation. EMBO J. 2008;27:1747–57.
Offer S, Menard JA, Perez JE, de Oliveira KG, Chandran VI, Johansson MC, et al. Extracellular lipid loading augments hypoxic paracrine signaling and promotes glioma angiogenesis and macrophage infiltration. J Exp Clin Cancer Res. 2019;38:241.
Zheng Y, Bao J, Zhao Q, Zhou T, Sun X. A spatio-temporal model of macrophage-mediated drug resistance in glioma immunotherapy. Mol Cancer Ther. 2018;17:814–24.
Bardi GT, Smith MA, Hood JL. Melanoma exosomes promote mixed M1 and M2 macrophage polarization. Cytokine. 2018;105:63–72.
Orecchioni M, Ghosheh Y, Pramod AB, Ley K. Macrophage polarization: different gene signatures in M1(LPS+) vs. classically and M2(LPS-) vs. alternatively activated macrophages. Front Immunol. 2019;10:1084.
Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8.
Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12.
Sasaki H, Zlatescu MC, Betensky RA, Ino Y, Cairncross JG, Louis DN. PTEN is a target of chromosome 10q loss in anaplastic oligodendrogliomas and PTEN alterations are associated with poor prognosis. Am J Pathol. 2001;159:359–67.
Han F, Hu R, Yang H, Liu J, Sui J, Xiang X, et al. PTEN gene mutations correlate to poor prognosis in glioma patients: a meta-analysis. Onco Targets Ther. 2016;9:3485–92.
Wei Q, Clarke L, Scheidenhelm DK, Qian B, Tong A, Sabha N, et al. High-grade glioma formation results from postnatal pten loss or mutant epidermal growth factor receptor expression in a transgenic mouse glioma model. Cancer Res. 2006;66:7429–37.
Hu X, Pandolfi PP, Li Y, Koutcher JA, Rosenblum M, Holland EC. mTOR promotes survival and astrocytic characteristics induced by Pten/AKT signaling in glioblastoma. Neoplasia. 2005;7:356–68.
Kwon CH, Zhao D, Chen J, Alcantara S, Li Y, Burns DK, et al. Pten haploinsufficiency accelerates formation of high-grade astrocytomas. Cancer Res. 2008;68:3286–94.
Hu JL, Wang W, Lan XL, Zeng ZC, Liang YS, Yan YR, et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol Cancer. 2019;18:91.
Zheng R, Du M, Wang X, Xu W, Liang J, Wang W, et al. Exosome-transmitted long non-coding RNA PTENP1 suppresses bladder cancer progression. Mol Cancer. 2018;17:143.
Xu B, Zhang Y, Du XF, Li J, Zi HX, Bu JW, et al. Neurons secrete miR-132-containing exosomes to regulate brain vascular integrity. Cell Res. 2017;27:882–97.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81672499, 81000963), Jiangsu Province's Natural Science Foundation (BK20211115), the Youth Medical Talent Project of the Jiangsu Province (QNRC2016470), Top Talent Project of "six one projects" for High-level Health talents in Jiangsu Province (LGY2018038) and Six talent peaks project in Jiangsu Province (YY-206), Jiangsu Postdoctoral Research Funding Program (2019K061) and China Postdoctoral Research Funding Program (2019M661906).
Author information
Authors and Affiliations
Contributions
LS and YC performed most of the experiments, treated data, and wrote the manuscript. WY performed mouse treatment experiments. GS participated in research conception and design, and revised the manuscript. JG supervised experiments and contributed to analytic tools. All authors read and approve the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The studies involving human participants were reviewed and approved by the Ethics Committee of The First people’s Hospital of Yancheng. The participants provided their written informed consent to participate in this study. All animal studies obtained the approval of the Institutional Animal Care and Use Committee of The First people’s Hospital of Yancheng and implemented in line with institutional and national guidelines (No. 2015-007).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Professor Stephen Tait
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shi, L., Cao, Y., Yuan, W. et al. Exosomal circRNA BTG2 derived from RBP-J overexpressed-macrophages inhibits glioma progression via miR-25-3p/PTEN. Cell Death Dis 13, 506 (2022). https://doi.org/10.1038/s41419-022-04908-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-022-04908-4
This article is cited by
-
Exosome derived from tumor-associated macrophages: biogenesis, functions, and therapeutic implications in human cancers
Biomarker Research (2023)
-
Exosomal circRNA: emerging insights into cancer progression and clinical application potential
Journal of Hematology & Oncology (2023)
-
Recent advances of exosomal circRNAs in cancer and their potential clinical applications
Journal of Translational Medicine (2023)
-
miRNA-seq identification and clinical validation of CD138+ and circulating miR-25 in treatment response of multiple myeloma
Journal of Translational Medicine (2023)